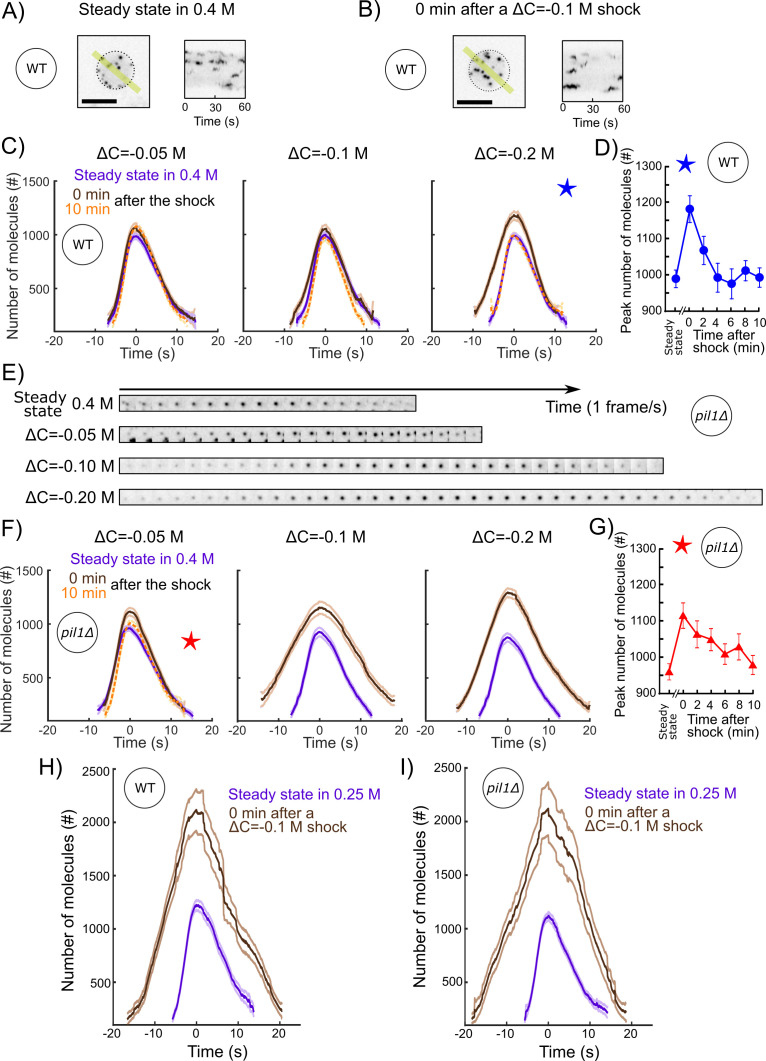
Figure 7. The actin endocytic machinery adapts to increases of membrane tension in protoplasts.
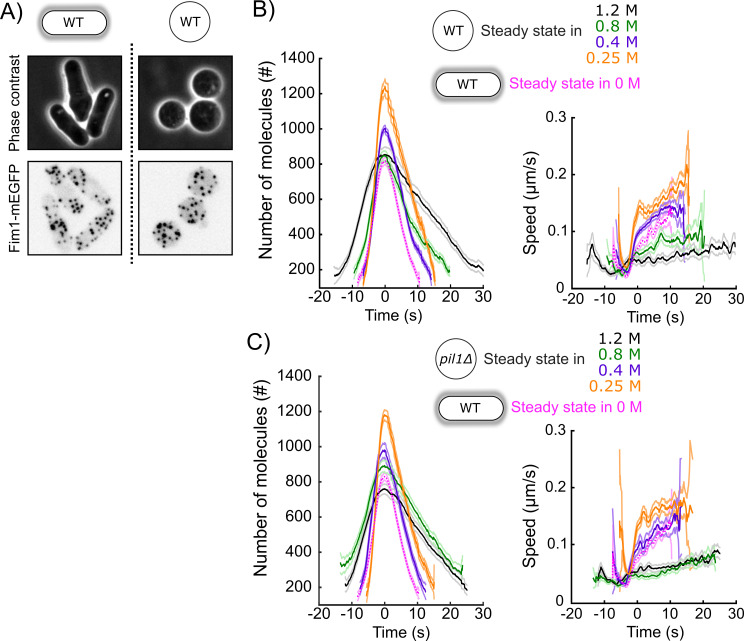
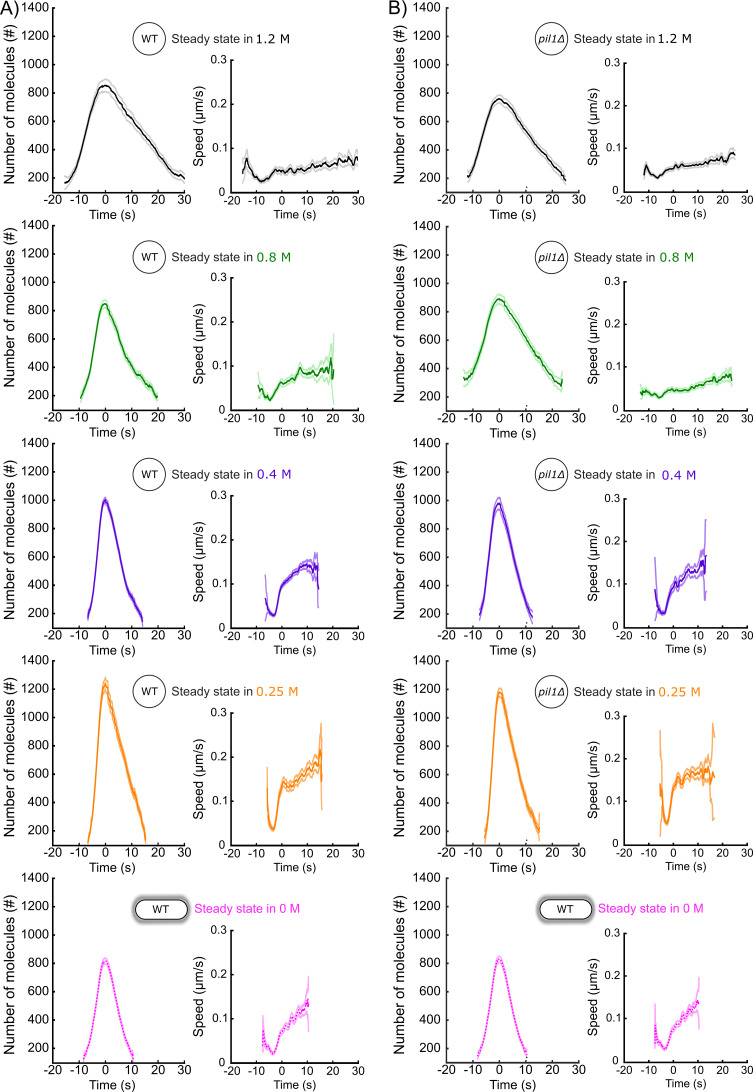
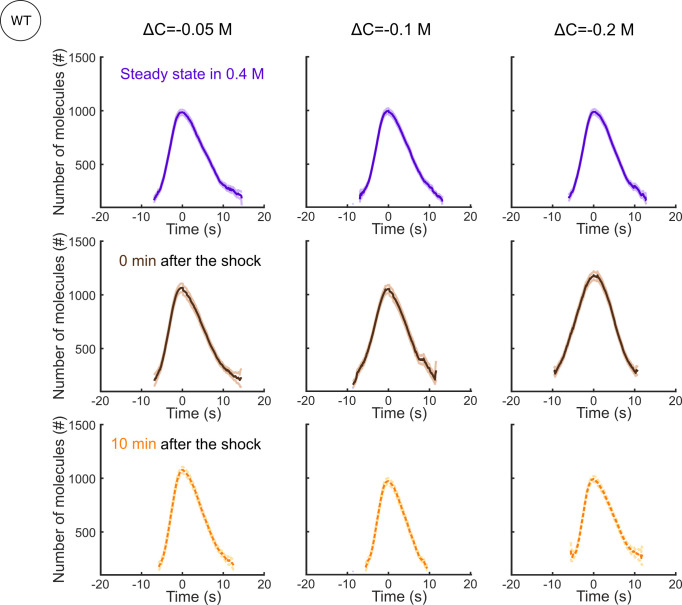
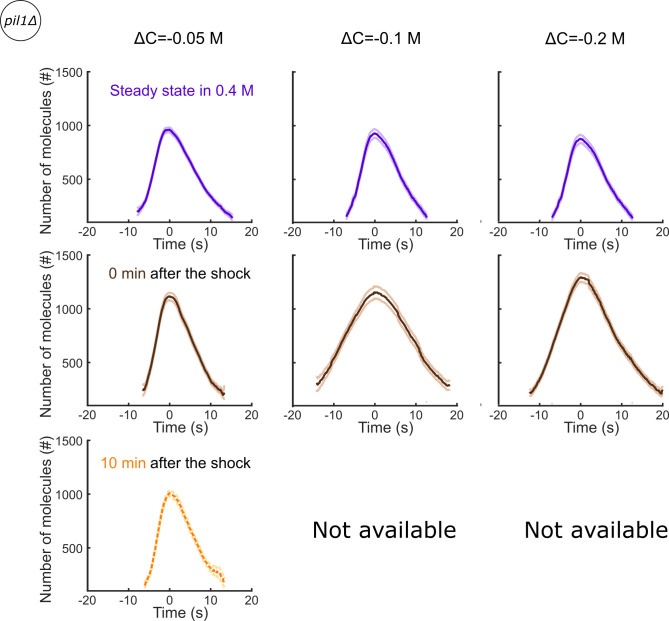
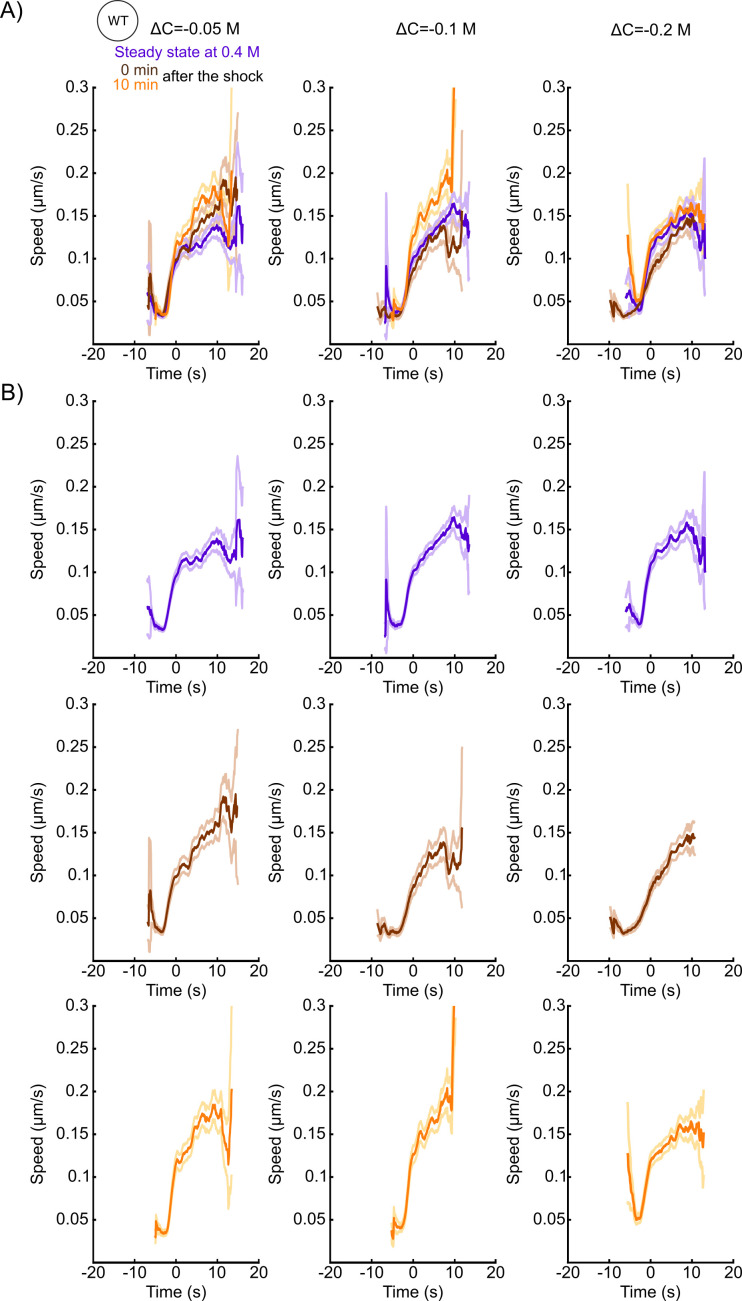
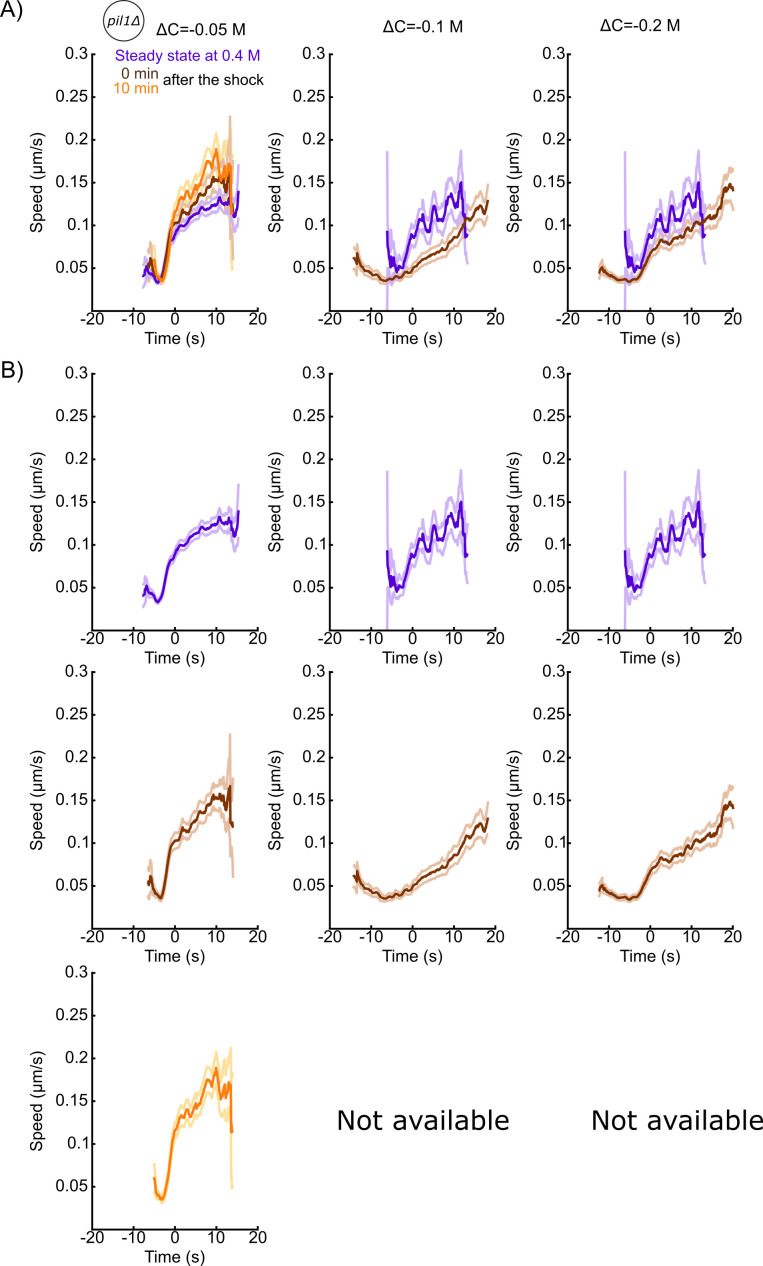
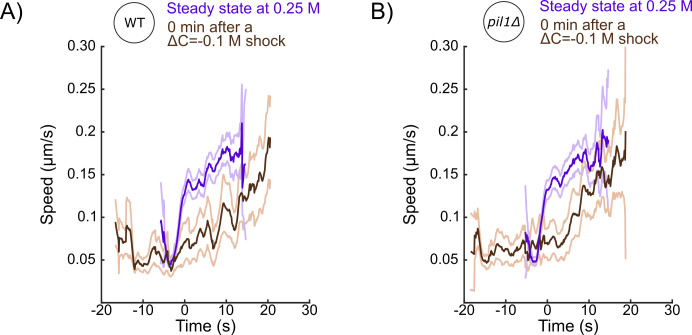
(A and B) Representative wild-type protoplasts expressing Fim1-mEGFP (inverted contrast) at steady state in 0.25 M sorbitol (A, left panel) and immediately after (0 min) an acute osmotic shock of ΔC = −0.1 M (B, left panel). Right panels: kymographs of the fluorescence under the yellow lines in the left panels. Black dashed lines: protoplast outline. Scale bars: 5 µm. (C and F) Number of Fim1p-mEGFP molecules in wild-type (C) and pil1Δ (F) protoplasts at steady state in 0.4 M sorbitol (purple), 0 min (brown) and 10 min (orange) after a hypotonic shock of ΔC = −0.05 M (left panels), ΔC = −0.1 M (middle panels), and ΔC = −0.2 M (right panels), N ≥ 95. Data for each condition are plotted separately in Figure 7—figure supplement 3 (wild type) and Figure 7—figure supplement 4 (pil1Δ). The speeds of Fim1p-mEGFP for each condition are shown in Figure 7—figure supplement 5 (wild type) and Figure 7—figure supplement 6 (pil1Δ). The numbers of endocytic events used in each curve are given in Supplementary file 1i. Note that the large majority of pil1Δ protoplasts were too damaged or dead 2 min after hypotonic shocks larger than or equal to ΔC = −0.1 M to allow us to track enough endocytic events and produce a curve (Figure 2B,C and Figure 7—figure supplement 1). In panel (C), the difference in the number of molecules at time 0 s at steady state and 0 min after the shock is statistically significant for all shocks (one-way ANOVA, p<0.03), and the difference between steady state and 10 min after the shock is not statistically significant (one-way ANOVA, p>0.2; details in the data file for C). In panel (F), the difference at steady state and 0 min after the shock is statistically significant for all shocks (one-way ANOVA, p<10−5; details in the data file for Figure 5F). (D) Temporal adaptation of the peak number of Fim1p-mEGFP in wild-type protoplasts initially at steady state in 0.4 M sorbitol and 0–10 min after a ΔC = −0.2 M osmotic shock. The condition for this figure is the same as the condition with the blue star in (C). The difference between steady state and 0 or 2 min after shock is statistically significant (one-way ANOVA, p<10−3; details in the data file for D). The difference between steady state and 4, 6, 8, and 10 min after shock is not statistically significant (one-way ANOVA, p>0.2; details in the data file for D). (E) Montage of representative endocytic events (Fim1-mEGFP, inverted contrast) in pil1Δ protoplasts (one frame per second) at steady state in 0.4 M sorbitol (first row) and immediately after (0 min) hypotonic shocks of ΔC = −0.05 M (second row), ΔC = −0.10 M (third row), and ΔC = −0.20 M (fourth row). (G) Temporal adaptation of the peak number of Fim1p-mEGFP in pil1Δ protoplasts initially at steady state in 0.4 M sorbitol and 0–10 min after a ΔC = −0.05 M shock. The condition in this figure is the same as the condition with the red star in (F). The difference between steady state and 0, 2, 4, 6, or 8 min after shock is statistically significant (one-way ANOVA, p<0.01; details in the data file for F). The difference between steady state and 10 min after shock is not statistically significant (one-way ANOVA, p>0.3; details in the data file for F). (D and G) Error bars are 95% confidence intervals. The numbers of endocytic events at each time point are given in Supplementary file 1j. (H and I) Number of molecules of Fim1p-mEGFP for wild-type (H) and pil1Δ (I) protoplasts at steady state in 0.25 M sorbitol (purple dashed) and immediately after (0 min) a hypotonic shock of ΔC = −0.1 M (brown), N ≥ 67. The difference in the number of molecules at time 0 s at steady state and 0 min after the shock is statistically significant for all conditions (one-way ANOVA, p<10−16). The speed data for each condition are plotted in Figure 7—figure supplement 7. The numbers of endocytic events used in each curve are given in Supplementary file 1k. The survival rates for the wild-type and pil1Δ protoplasts in these conditions are plotted in Figure 2D.