Abstract
COVID-19 was caused by a novel coronavirus known as SARS-CoV-2. The COVID-19 disease outbreak has been avowed as a global pandemic by the World Health Organization at the end of March 2020. It leads to the global economic crash, resulting in the starvation of a large population belonging to economically backward countries. Hence, the development of an alternative medicine along with the vaccine is of the utmost importance for the management of COVID-19. Therefore, screening of several herbal leads was performed to explore their potential against SARS-CoV-2. Furthermore, viral main protease was selected as a key enzyme for performing the study. Various computational approaches, including molecular docking simulation, were used in the current study to find potential inhibitors of viral main protease from a library of 150 herbal leads. Toxicity and ADME prediction of selected molecules were also analysed by Osiris molecular property explorer software. Molecular dynamic simulation of the top 10 docked herbal leads was analysed for stability using 100 ns. Taraxerol (−10.17 kcal/mol), diosgenin (10.12 kcal/mol), amyrin (−9.56 kcal/mol), and asiaticoside (−9.54 kcal/mol) were among the top four herbal leads with the highest binding affinity with the main protease enzyme. Thus, taraxerol was found to be an effective drug candidate against the main protease enzyme for the management of COVID-19. Furthermore, its clinical effect and safety profile need to be established through an in vivo model.
Supplementary information
The online version contains supplementary material available at 10.1007/s11224-022-01943-x.
Keywords: Taraxerol, SARS-CoV-2, COVID-19, Herbal leads, Main protease, Molecular docking
Introduction
The coronavirus disease (COVID-19) outbreak was caused by SARS-CoV-2 and spread from Wuhan, China, at the end of December 2019 [1, 2]. COVID-19 was thought to have emerged from a seafood market from an unknown species, causing the virus’s emergence and symptoms to be comparable with pneumonia [1]. The confirmation of human-to-human transmission of this contagious virus was made by the National Health Commission of China in late January 2020. As SARS-CoV-2 began to spread across international borders, affecting the populations of multiple countries, the WHO designated it as an internationally concerning public health emergency and declared it a global pandemic in March 2020 [3].
Based on the sequence resemblance of the SARS-CoV-2 nucleotide, it is considered a betacoronavirus, having a high similarity to well-known and aggressive strains of human coronaviruses (HCOVs). The nuclear material of SARS-CoV-2 was found to be a long positive-sense single-stranded RNA encoding two discrete types of structural as well as non-structural viral proteins [1, 2]. The 5′-untranslated region (5′UTR) and a replicase complex encode for viral structural proteins like spike, nucleocapsid, matrix, and envelope, while the 3′UTR and several unidentified open reading frames encode for viral non-structural proteins like protease, phosphatase, and polymerases [1, 2].
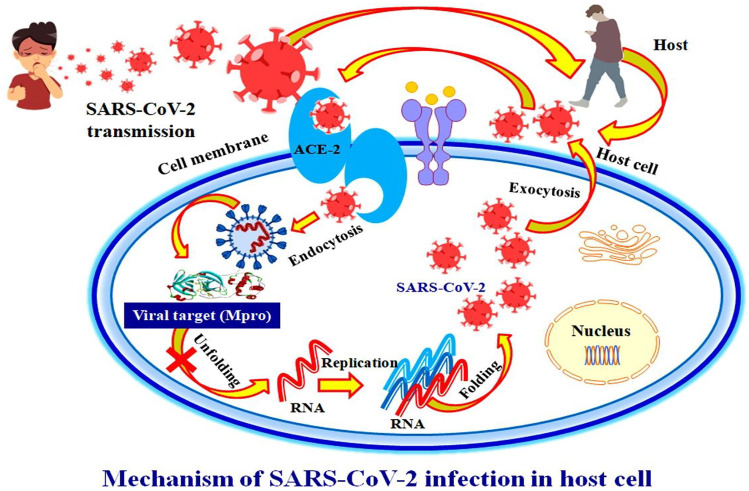
COVID-19 has initial symptoms of cough, malaise, fatigue, fever, body ache, loss of smell or taste sensation, inadequacy, and shortness of breath followed by respiratory distress [4]. Persistent viral infection affects not only the lungs but also other key organs of the body, eventually leading to organ failure [5–7]. Coronavirus transmits from person to person through coughing and sneezing, which spreads through the nasal mucosa via airborne droplets, where it replicates narrowly in ciliated epithelial cells, causing cell damage and inflammation. The transmission of this virus was also observed from infected surfaces to an individual’s hands and then to their bodies [8].
Globally, more than 216 countries have been affected by the pandemic outbreak of COVID-19 until December 31, 2021. More than 287,022,026 confirmed cases of coronavirus-affected people have been reported, with more than 5,447,886 deaths. According to the WHO, China is the first country to report 102,083 confirmed cases of COVID-19 with 4636 mortalities. In addition, new cases of COVID-19 are being reported day by day, in the USA, with more than 55 million confirmed cases and 845,745 deaths, followed by India with more than 34 million confirmed cases and Brazil with more than 22 million confirmed cases [9]. Even after 18 months of the outbreak of the novel SARS-CoV-2, there is still no approved therapy or vaccine available for the cure of this lethal infectious disease. In light of the foregoing, it is critical to design a novel remedy to combat this worldwide epidemic crisis.
The role of viral main protease enzyme has been identified in causing infection. It plays an important role in pathogenic entry into the host via ACE2 and could be used as a therapeutic antiviral drug target. Inhibiting the viral main protease restricts the pathogen’s entry into the host cell and terminates the viral infection [4, 10]. As a result, an inhibitor of the main protease is considered to be a successful treatment approach for COVID-19 pandemics all over the world. The proven methodology for in silico screening of herbal leads to identify modest inhibitors of the viral main protease enzyme is depicted graphically in Fig. 1.
Fig. 1.
An in silico approach for screening herbal leads for potential inhibitors of the viral main protease enzyme in order to discover new antiviral therapies against SARS-CoV-2
Herbal medicine is a holistic medicine, having being used for the management of several health problems for thousands of years. Herbal medicine inspired several drugs, such as artemisinin, paclitaxel, reserpine, morphine, quinine, emetine, aspirin, and many more, which have been discovered for the treatment of numerous diseases [11]. Most of the world’s population relies on herbal medicine as an alternative and complementary medicine for the management of diseases like COVID-19. In this context, several herbal medicines and their bioactive leads including Camellia sinensis (epigallocatechin gallate), Andrographis paniculata (andrographolide), Artemisia annua (artemisinin), Betula sp. (betulinic acid), Citrus sp. (hesperidin), Curcuma longa (curcumin), Ficus benjamina (biorobin), Glycyrrhiza glabra (glycyrrhizin), Mollugo cerviana (vitexin), Myristica fragrans (myricitrin), Piper nigrum (piperine), Radix sophorae (matrine), Stephania tetrandra (tetrandrine), Tinospora cordifolia (berberine), Torreya nucifera (luteolin), etc. have been explored through computational approaches for the treatment of COVID-19 [12–32]. Hence, it inspires us to find such herbal leads through an in silico computational approach for the management of the current pandemic situation. Thus, in the current investigation, putative antagonists of the main protease enzyme of the SARS-CoV-2 were acknowledged using docking-based computational screening of herbal ligands. Moreover, in order to develop novel therapy to combat COVID-19, validation of herbal leads with respect to time by using molecular dynamics (MD) simulation was done.
Material and methods
Preparation of compound library
A molecular ligand library was prepared by considering the diverse compounds already reported for their antiviral properties [33–38]. The already reported antiviral compounds were used to prepare a ligand library in the current study with the intention of validating their effectivity against the SARS-CoV-2 as well as establishing the probable mechanism of action of those compounds which are supposed to be interacting with the main protease enzyme for their antiviral effect against SARS-CoV-2.
Molecular docking simulation-based virtual screening
The main protease enzyme of the SARS-CoV-2 complex with an antagonist N3 was procured from the protein databank (PDBID: 6lu7) [10, 39, 40]. The parted macromolecular model was set for docking by amputation of superfluous water molecules and the addition of polar hydrogens, while the reference ligand N3 was prepared by assigning rotatable, non-rotatable, and un-rotatable bonds [41, 42]. The interacting residues of the viral enzyme with the complex inhibitor were evaluated by using PyMol software to confirm the macromolecular active site and prepare a grid box [42, 43]. The prepared grid box was further utilised by the Autogrid module to evaluate the chemical configuration of the macromolecule as well as the ligand to create maps for various atoms, necessary to accomplish docking simulation [41, 42].
The confirmational search by Autodock for the execution of the docking process was performed by the Lamarckian Genetic Algorithm (LGA). The force-field calculates the ligand’s binding energy by integrating intramolecular energies and assessing energetics for bound and unbound states using a comprehensive thermodynamic model. Each ligand molecule’s docking parameters is stored in the docking parameter file (DPF) [44, 45].
For validation purposes, the reference ligand N3 was docked against the macromolecular target. Validation of the grid parameters as well as protocols used in the current docking procedure was performed by considering the chemical interactions as well as the conformation of the docked ligand N3 with respect to its crystallised bioactive conformation.
Validated docking protocols were further utilised for computational screening of a ligand library consisting of 150 diverse herbal ligands against the viral main protease. Virtual screening of herbal ligands was performed with the intent of identifying prospective lead compounds targeting the viral main protease enzyme [42].
The interactions of the ligands against the macromolecular residues were taken into account while evaluating the docking outcomes for all ligands screened against the viral main protease enzyme. The compounds with the minimum binding energy were selected as the lead molecules based upon the predefined empirical range for the obtained binding energy of −5 to −15 kcal/mol [46–50].
Prediction of pharmacokinetics and toxicological properties
Evaluation of physicochemical, pharmacokinetics, and toxicological parameters of the lead molecules obtained after the docking-based screening were predicted by using the pkCSM-pharmacokinetics server. The online server screens the ligand molecule for smooth kinetics within the body required for its physiological expression without toxicity [51].
Lead molecules had their molecular weight (MW), partition coefficient (LogP), hydrogen bond acceptor/donor (HBA/HBD), rotatable bonds count, topological polar surface area (TPSA), and solubility (LogS) calculated. These parameters regulate the ADMET of the lead candidates. The permeability across the blood–brain barrier (BBB), gastrointestinal absorption, and the inhibitory potential of the cytochrome P450 isoenzymes were also considered for the leads to determine their druggability. These parameters were standardised with the Veber’s rule, the Lipinski’s rules of 5, and druggability [52, 53].
Molecular dynamic simulation
Amyrin, diosgenin, and taraxerol were further selected as potential leads of the viral main protease for further simulation to study the stability of their macromolecular complex with respect to time based on their docking score, observed chemical interactions against the target receptor, pharmacokinetic evaluation, as well as toxicity profiling. MD simulation for a shorter duration of timeframe, lasting 10 ns, was performed initially for all the three shortlisted lead molecules. Based on the results obtained for the 10-ns MD simulation, taraxerol was selected to further magnify the MD simulation for a longer time period of 100 ns.
The protein–ligand macromolecular complex was simulated for 100 ns by using Schrodinger’s Desmond module at a constant temperature of 300 K [54, 55]. MD simulations were performed by solvating the macromolecular complex in an explicit water box of size 10 and modelling the protein–ligand macromolecular complex using the OPLS3e force field [56–58]. Previously, promising and repeatable results were reported using the OPLS3e force field and the SPC water model for the protein targets complexed with the organic ligands [59]. By introducing the ions and subsequently minimising their energy, the macromolecular complex was neutralised. The Nose–Hoover thermostatic algorithm [60] was used to keep the energy-minimised macromolecular complex at 300 K, while the Martina–Tobias–Klein approach was used to keep the pressure constant throughout the simulations [61]. The long-range electrostatic interactions between the ligand and the macromolecule were calculated using the particle mesh Ewald (PME) [62] technique with 0.8 grid spacing and a cutoff radius of 9.0 for Coulomb interactions after the NPT-ensemble MD simulation was performed for 100 ns. The simulation interaction diagram tool in the Desmond was used to examine the ligand’s precise binding interactions with the macromolecular main protease [54, 55].
The root mean square deviation (RMSD) of the atoms of both receptors and the complexed ligand were calculated by comparing their reference frame to determine atomic displacement for a certain timeframe during the complexation process. During the simulation phase, the macromolecular residues’ root mean square fluctuation (RMSF) was calculated in reference to their initial condition in the crystalline structure. Throughout the simulation technique, the residue index was used to plot the distribution of macromolecular secondary structural elements (SSE) such as α-helices and β-strands. The binding interactions of the ligand within the macromolecular site were separated into four groups throughout the simulation procedure: hydrogen bonds, hydrophobic interactions, ionic interactions, and water interaction bridges. The radius of gyration (rGyR), molecular surface area (MolSA), solvent accessible surface area (SASA), and polar surface area of the ligands were all measured (PSA) [63–65]. In comparison to its initial frame, the RMSD value of the ligand molecule was determined throughout the whole simulation duration. The PSA was determined by taking into consideration the total contribution of oxygen and nitrogen atoms, whereas the ligand’s extended length, which correlates to its main moment of inertia, was computed using a 1.4 probe radius.
Results
Molecular docking simulation-based virtual screening
A three-dimensional structural model of the viral macromolecule with a resolution of 2.16 was obtained using the X-ray diffraction technique. Viral protein is made up of a single chain of 306 amino acid residues. Complex ligand N3 comprises 26 rotatable bonds and 9 aromatic carbons, making it a complex inhibitor. In the current computational investigation, all bonds of the inhibitor molecule were kept flexible, and it was also stored in the AutoDock software’s pdbqt format.
The ligand N3 and active binding residues of the viral main protease were wrapped together to prepare a three-dimensional grid box. The exact grid coordinates were obtained from our previously published study on the same protein [46–50, 63, 66]. The docking results for the complex inhibitor N3 against viral macromolecular targets are tabulated in Table 1.
Table 1.
Docking results of ligand N3 against the viral main protease enzyme
| Macromolecule | RMSD (Å) | Binding energy (kcal/mol) | Ki (nM) |
|---|---|---|---|
| 6lu7 | 0.88 | −8.22 | 935.62 |
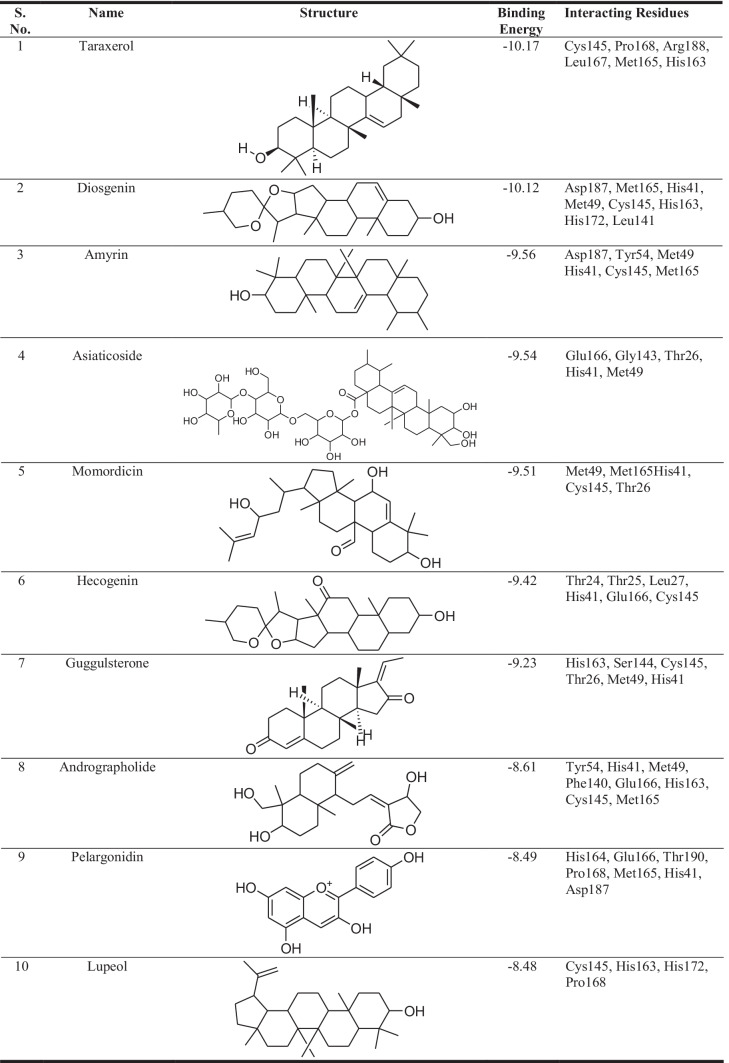
The lead molecules were chosen based on their affinity for the viral main protease enzyme. The lowest binding energy obtained for preeminent pose for each ligand and their binding interactions with the macromolecule were used to determine the binding affinity of the leads. Table 2 shows the binding energy acquired from docking-based virtual screening of the top 10 shortlisted herbal leads. The interacting residues were explored by analysing the binding site of the target macromolecule with the help of Discovery Studio visualizer software with the intent of identifying the important amino acid residues of the viral main protease enzyme.
Table 2.
Binding energy of short-listed herbal leads for the viral main protease enzyme
Prediction of pharmacokinetics and toxicological properties
The movement of drugs within the human body is regulated by physicochemical and pharmacokinetic parameters. These regulatory parameters are crucial for the pharmacological expression via pharmacodynamics as well as toxicological properties of a drug molecule. The pkCSM webserver was used for prediction of physicochemical, pharmacokinetic, as well as toxicological parameters of the lead molecules [51]. The physicochemical, pharmacokinetics, and pharmacodynamics properties of the selected lead molecules are tabulated in Table 3.
Table 3.
Physicochemical, pharmacokinetics, and pharmacodynamics properties of the selected lead molecules for the SARS-CoV-2 main protease enzyme
| Property | Descriptor | Taraxerol | Diosgenin | Amyrin | Asiaticoside | Momordicin | Hecogenin | Guggulsterone | Andrographolide | Pelargonidin | Lupeol |
|---|---|---|---|---|---|---|---|---|---|---|---|
| MW | (g/mol) | 412 | 414.63 | 426.73 | 959.13 | 472.71 | 430.63 | 312.45 | 350.45 | 271.25 | 426.73 |
| LogP | - | 7.78 | 5.71 | 8.025 | −1.0328 | 5.456 | 4.973 | 4.644 | 1.963 | 3.203 | 8.025 |
| Rotatable bond | - | 0 | 0 | 0 | 9 | 5 | 0 | 0 | 3 | 1 | 1 |
| HBA | - | 1 | 3 | 1 | 19 | 4 | 4 | 2 | 5 | 4 | 1 |
| HBD | - | 1 | 1 | 1 | 12 | 3 | 1 | 0 | 3 | 4 | 1 |
| TPSA | (Å)2 | 20.93 | 38.69 | 20.23 | 315.2 | 77.76 | 55.76 | 34.14 | 86.99 | 90.15 | 20.23 |
| Absorption | Water solubility (mol/L) | −6.628 | −5.684 | −6.906 | −2.797 | −4.759 | −5.453 | −5.401 | −3.045 | −3.393 | −5.958 |
| Absorption | Caco2 permeability | 1.299 | 1.227 | 1.358 | −0.637 | 0.951 | 1.186 | 1.477 | 1.213 | 0.37 | 1.356 |
| Absorption | Intestinal absorption (%) (human) | 97.705 | 96.491 | 97.891 | 5.812 | 97.953 | 95.816 | 100 | 94.961 | 85.871 | 99.707 |
| Absorption | Skin permeability (Log Kp) | −2.706 | −3.449 | −2.828 | −2.735 | −2.546 | −3.726 | −2.768 | −3.158 | −2.735 | −2.675 |
| Absorption | P-glycoprotein substrate | No | No | No | Yes | Yes | No | No | Yes | Yes | Yes |
| Absorption | P-glycoprotein I inhibitor | Yes | Yes | Yes | Yes | Yes | Yes | Yes | No | No | Yes |
| Absorption | P-glycoprotein II inhibitor | Yes | Yes | Yes | No | Yes | Yes | No | No | No | Yes |
| Distribution | VDss (human) | 0.157 | 0.52 | 0.321 | −0.364 | −0.277 | 0.272 | 0.343 | −0.289 | 0.17 | −0.177 |
| Distribution | Fraction unbound (human) | 0 | 0 | 0 | 0.373 | 0.021 | 0.001 | 0 | 0.225 | 0.125 | 0 |
| Distribution | BBB permeability | 0.724 | 0.298 | 0.713 | −2.675 | −0.467 | −0.17 | 0.19 | −0.766 | −1.221 | 0.721 |
| Distribution | CNS permeability | −1.419 | −2.903 | −1.925 | −5.015 | −1.847 | −1.484 | −2.021 | −2.484 | −2.088 | −1.261 |
| Metabolism | CYP2D6 substrate | No | No | No | No | No | No | No | No | No | No |
| Metabolism | CYP3A4 substrate | Yes | Yes | Yes | No | No | Yes | Yes | Yes | No | Yes |
| Metabolism | CYP1A2 inhibitor | No | No | No | No | No | No | No | No | Yes | No |
| Metabolism | CYP2C19 inhibitor | No | No | No | No | No | No | No | No | Yes | No |
| Metabolism | CYP2C9 inhibitor | No | No | No | No | No | No | No | No | Yes | No |
| Metabolism | CYP2D6 inhibitor | No | No | No | No | No | No | No | No | No | No |
| Metabolism | CYP3A4 inhibitor | No | No | No | No | No | No | Yes | No | Yes | No |
| Excretion | Total clearance (log ml/min/kg) | 0.004 | 0.328 | 0.119 | 0.286 | 0.457 | 0.311 | 0.61 | 1.183 | 0.692 | 0.153 |
| Excretion | Renal OCT2 substrate | No | Yes | No | No | No | Yes | No | No | No | No |
| Toxicity | AMES toxicity | No | No | No | No | No | No | No | No | No | No |
| Toxicity | Max. tolerated dose (human) (log mg/kg/day) | −0.194 | −0.234 | −0.275 | −0.847 | −1.438 | −0.588 | −0.355 | −1.1 | 0.787 | −0.263 |
| Toxicity | hERG I inhibitor | No | No | No | No | No | No | No | No | No | No |
| Toxicity | hERG II inhibitor | Yes | No | Yes | Yes | No | No | Yes | No | No | No |
| Toxicity | Oral rat acute toxicity (LD50) (mol/kg) | 2.861 | 1.812 | 2.345 | 2.516 | 4.003 | 1.927 | 1.801 | 2.786 | 2.436 | 3.215 |
| Toxicity | Oral rat chronic toxicity (LOAEL) (mg/kg/day) | 1.144 | 1.562 | 1.031 | 8.049 | 1.765 | 1.294 | 1.78 | 0.389 | 1.595 | 1.688 |
| Toxicity | Hepatotoxicity | No | No | No | No | Yes | No | Yes | No | No | No |
| Toxicity | Skin sensitisation | No | No | No | No | No | No | No | No | No | No |
| Toxicity | T. Pyriformis toxicity (mg/L) | 0.394 | 0.397 | 0.418 | 0.285 | 0.364 | 0.376 | 1.014 | 0.346 | 0.435 | 0.335 |
| Toxicity | Minnow toxicity | −1.552 | −0.229 | −1.953 | 10.986 | 0.095 | 0.563 | −0.028 | 1.157 | 0.957 | −2.002 |
Taraxerol has all the parameters well within the predefined range according to Lipinski’s rule of five, i.e., MW < 500, < 10 HBA, < 5 HBD, TPSA 20–130 Å2, except LogP value, which exceeds the limit of 5.0. The demonstrated physicochemical properties suggest that taraxerol has optimised pharmacokinetics for drug-like candidature [51]. Taraxerol has not displayed a substrate-like expression for P-glycoprotein, which acts as a biological barrier for xenobiotics and exogenous toxins.
Taraxerol has not shown substrate-like expression for most of the cytochrome P450 isoenzymes, except for the metabolic CYP3A4. The ligand consistently exhibits a high proclivity for excretion from biological systems and low expression across a variety of toxicity pathways such as AMES, hERG I inhibition, hepatotoxicity, skin sensitization, Tetrahymena pyriformis toxicity, and minnow toxicity. Cumulatively, the predicted physicochemical, pharmacokinetics, pharmacodynamics, and toxicological parameters for taraxerol fall well within the predefined range, clearly indicating its promising potential as a druggable candidate [51–53].
Molecular dynamic simulation
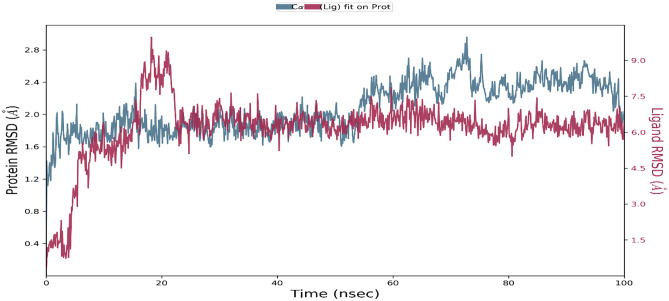
The MD simulation of the viral main protease enzyme for a shorter timeframe of 10 ns found taraxerol to be the most stabilised inhibitor molecule complex in the active cavity of the viral main protease enzyme. The putative herbal inhibitor compound taraxerol was further validated by using the Schrodinger’s Desmond module to run a 100-ns MD simulation. There are 306 residues in the macromolecular receptor, but the ligand atom contains only one rotatable bond and 30 heavy atoms out of a total of 78. Based on structural validation throughout the operation, the RMSD analysis supports the smooth execution of the equilibrated simulation process. The ligand RMSD demonstrates the ligand’s stability in relation to the macromolecular binding residues during the simulation procedure by aligning their heavy metals.
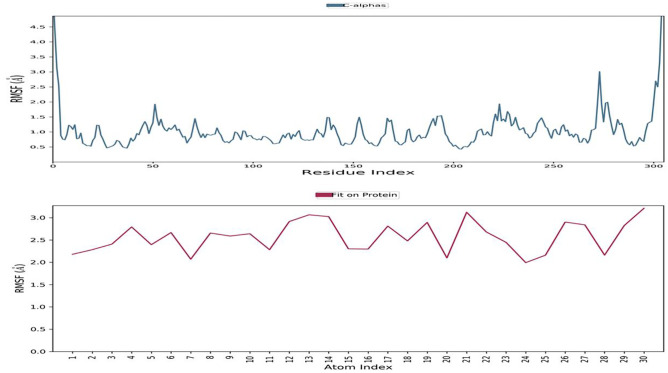
The RMSD value for the macromolecular residues was found to be well within the 2.8 Å range, indicating that the majority of the residues do not shift from their starting position during ligand molecule complexation. The ligand molecule’s RMSD value has been maintained well between 6 and 7.5 Å throughout the simulation run, despite some early swings of up to 9.0 Å during ligand adjustment within the macromolecular binding cavity, indicating the ligand’s adjustment within the macromolecular cavity during the initial 20 ns of the simulation, followed by achieving its stable conformation for the remaining 80-ns duration of the simulation process. The ligand taraxerol undergoes a sequence of vibrations after reaching the active binding site of the viral main protease to achieve the most stable confirmation inside the active binding site. As a result, the early oscillations in the ligand’s RMSD value during 3–10 ns are caused by these continuing vibrations while performing certain manoeuvres to achieve the most stable confirmation inside the active binding site of the viral main protease enzyme. Figure 2 shows the RMSD of the protein and ligand over the 100-ns period of the MD simulation. The RMSF value of macromolecular residues was determined to be well within the allowed range of 3 Å. A few residues changed slightly, with an RMSF value of 2–3 Å. However, the bulk of residues exhibited smaller variations, with an average value of less than 1 Å. Figure 3a, b show the RMSF of the viral main protease and the complex ligand taraxerol during the 100-ns period of the MD simulation. The RMSF observed for the ligand taraxerol complex within the active binding site of the viral main protease enzyme was found to be within the range of 2–3 Å throughout the 100-ns timeframe. This clearly suggests that the ligand was stabilised within the active site of the target macromolecule with slight fluctuations of some functionals required for interacting with the target macromolecule.
Fig. 2.
Root mean square deviation (RMSD) of the viral main protease enzyme and ligand taraxerol observed during the 100-ns MD simulation
Fig. 3.
Root mean square fluctuation: During the 100-ns timeframe of the MD simulation, the RMSF of the viral main protease enzyme and the complexed ligand taraxerol were measured
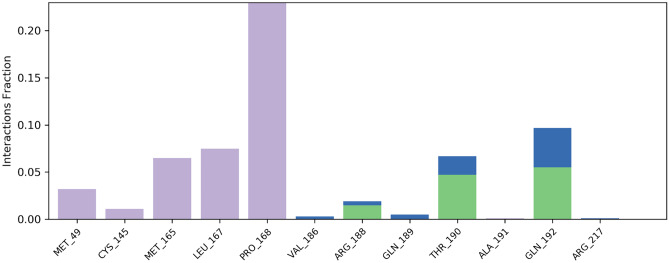
During the simulation process, the SSE analysis revealed that it has 14.94% alpha helices and 24.21% beta-sheets, resulting in a total contribution of 39.14% SSE, which may be conserved for the majority of the simulation. The macromolecular ligand interaction study revealed that Pro168, Leu167, Met165, Cys145, Met49, Gln192, Thr190, and Arg188 interact with the ligand over the course of the 100-ns simulation procedure. More than eight macromolecular residues were discovered to be consistently interacting with the complex ligand throughout the simulation run. Figure 4 depicts the detailed protein–ligand connections observed during the whole timeframe of the 100-ns MD simulation. The ligand’s RMSD value was substantially within the range of 1.5 Å, indicating that the ligand fluctuated as little as possible during the simulation process. The ligand’s rGyr value was discovered to be in the range of 4.1–4.3 Å. During the modelling phase, no intramolecular hydrogen bonding in the ligand was observed. Throughout the modelling phase, the ligand’s MolSA was discovered to be in the range of 372–384Å2. After some initial variations, the ligand’s SASA was found to be in the range of 150–300Å2 during the simulation process. The PSA for the complex ligand was discovered to be between 35 and 40 Å2 during the simulation process.
Fig. 4.
Protein–ligand contacts: Detailed protein–ligand interactions were found during the 100-ns MD simulation timescale
Discussion
The in silico computational approach is pioneering for drug discovery and development. Several drugs have been identified for the treatment of many diseases, including viral infections. There are several examples of in silico inspired medicines such as chloroquine, hydroxychloroquine, remdesivir, ritonavir, lopinavir, darunavir, arbidol, peptide EK1, cobicistat, amodiaquine, niclosamide, telbivudine, nicotinamide, ribavirin, sofosbuvir, galidesivir, tenofovir, setrobuvir, artemisinin, mefloquine, halofantrine, lumefantrine, amodiaquine, primaquine, doxycycline, atovaquone, sulfonamides didanosine, camptothecin, teicoplanin, velpatasvir, ledipasvir, and favipiravir that have been explored against COVID-19. But their clinical uses are limited due to quality-related safety issues. Most of the drugs are under clinical trials [33].
As we know, the world is facing health and economic problems due to COVID-19. Still, there are no such therapies as some vaccines have been approved for the treatment of COVID-19. There is a continuous quest to find drugs for treating COVID-19. In this context, herbal medicine is very popular throughout the globe for its miraculous effect on health problems. It has been used as an alternative medicine for treating various diseases. As per the WHO, 80 percent of the world’s population depends upon herbal medicine for the cure and treatment of several diseases. Several drugs have been discovered through natural resources, including plants [67]. Therefore, it occupies our mind to identify such potential herbal leads for the management of COVID-19. In the recent past, numerous herbal leads like luteolin, psoralidin, caffeic acid, myricetin, quercetin, vitexin, tryptanthrin, shikonin, silvestrol, scutellarein, biorobin, hesperidin, andrographolide, catechins, reserpine, emetine, artemisinin, quinine, paclitaxel, and morphine have been recognised through computational approaches. They have exhibited remarkable inhibitory activity against SARS-CoV-2. Various research studies at preclinical level (in vitro and in vivo) and clinical trials are going on to ensure their therapeutic efficacy and safety [33].
In the present study, lead molecules with binding energy such as taraxerol (−10.17 kcal/mol), diosgenin (−10.12 kcal/mol), amyrin (−9.56 kcal/mol), asiaticoside (−9.54 kcal/mol), momordicin (−9.51 kcal/mol), hecogenin (−9.42 kcal/mol), guggulsterone (−9.23 kcal/mol), andrographolide (−8.61 kcal/mol), pelargonidin (−8.49 kcal/mol), and lupeol (−8.48 kcal/mol) were identified for inhibiting the viral main protease enzyme. Based on the observed chemical interactions, which are tabulated in Table 2, among the shortlisted leads against the viral macromolecular targets, a SAR was developed that confirms the involvement of macromolecular residues such as Cys145, Pro168, Met165, Leu167, Gln192, Thr190, and Met49 in the active ligand binding. These leads have been reported in medicinal plants, including Clitoria ternatea (taraxerol), Dioscorea sp. (diosgenin), Eclipta alba (amyrin), Centella asiatica (asiaticoside), Momordica charantia (momordicin), Chlorophytum borivilianum (hecogenin), Commiphora sp. (guggulsterone), Andrographis paniculata (andrographolide), Anagallis monelli (pelargonidin), and Carissa spinarum (lupeol) [11]. As a result, these bioactive-containing medicinal plants would be effective in managing the SARS-CoV-2 health issue. In another study, Kar et al. reported the inhibitory potential of Clerodendrum trichotomum containing lead taraxerol, friedelin, and stigmasterol against SARS-CoV-2 through an in silico approach [68].
Taraxerol, among other things, inhibited SARS-CoV-2 enzymes (spike protein (6lzg) [−7.5 0.01 kcal/mol]), main protease (8.4 0.01 kcal/mol), and RNA-dependent RNA polymerase (RdRp) [−7.4 0.02 kcal/mol] [69]. Taraxerol has been reported for several biological activities, including antiplasmodial (Plasmodium falciparum, IC50, 8.5 µM), antiparasitic (Trypanosoma brucei, IC50, 10.5 µM), antioxidant (IC50, 500 µM), etc. [70].
Similarly, Enmozhi et al. have reported the antiviral activity of andrographolide on SARS-CoV-2 main protease enzyme through in silico investigation. This investigation includes molecular docking, target analysis, toxicity prediction, and ADME prediction for andrographolide. Andrographolide, a bioactive component of A. paniculata, showed an inhibitory effect with binding efficiency (−3.094357 kcal/mol) against the main protease enzyme [13, 71]. Moreover, other lead compounds from plants (epicatechin gallate −7.24 kcal/mol, catechin −7.05 kcal/mol, kaempferol −9.41 kcal/mol, gingerol −6.67 kcal/mol, quercetin −8.58 kcal/mol, curcumin −7.31 kcal/mol, and demethoxycurcumin −8.17 kcal/mol) have shown inhibitory effects against the main protease enzyme [72]. In addition, phytoconstituents like boldine, ginkgetin, isoboldine, laurolitsine, lauroscholtzine, laurotetanine, norisoboldine, pseudolycorine, secoboldine, and syringic acid have shown good inhibitory potential against the main protease enzyme through in silico studies. The best binding affinity has been produced by laurolitsine as compared to other molecules [73]. Further, efficacy, drug/herb-drug interaction, safety, and toxicity profile of taraxerol need to be validated against SARS-CoV-2 through preclinical and clinical investigation. Hence, an in silico computational approach could be promising for getting new herbal leads for the management of COVID-19. Other alternate treatment options such as stem cells, monoclonal antibodies, plasma, and polypeptides are also under investigation for COVID-19.
Conclusion
The in silico virtual screening technique is a highly adequate, prudent, and quick approach for identifying a potent herbal lead having therapeutic activity against the main protease enzyme of COVID-19. Taraxerol was found to be a potential lead against the main protease enzyme of SARS-CoV-2. The binding site analysis has revealed that the residues Cys145, Pro168, Met165, Leu167, Gln192, Thr190, and Met49 play an important interacting role in the ligand binding. The results obtained by molecular docking simulation were further validated by performing 100-ns molecular dynamics simulation, and the pharmacokinetic profiling based on their physicochemical parameters was also performed to strengthen the candidature of taraxerol as a safe and optimised inhibitor of the viral main protease enzyme. Further, this herbal molecule needs to be validated through preclinical and clinical investigation for its therapeutic applicability.
Supplementary information
Below is the link to the electronic supplementary material.
Author contribution
Both the authors have contributed equally for the execution of current research. The framework is designed and executed by SM, while the validation of the research outcomes and drafting of the manuscript was done by RH.
Data availability
Yes.
Declarations
Conflict of interest
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Somdutt Mujwar, Email: somduttmujwar@gmail.com.
Ranjit K. Harwansh, Email: harwanshranjeet@gmail.com
References
- 1.Elfiky AA. Anti-HCV, nucleotide inhibitors, repurposing against COVID-19. Life Sci. 2020;248:117477. doi: 10.1016/j.lfs.2020.117477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Elfiky AA. Ribavirin, remdesivir, sofosbuvir, galidesivir, and tenofovir against SARS-CoV-2 RNA dependent RNA polymerase (RdRp): a molecular docking study. Life Sci. 2020;253:117592. doi: 10.1016/j.lfs.2020.117592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sohrabi C, Alsafi Z, O'Neill N, Khan M, Kerwan A, Al-Jabir A, Iosifidis C, Agha R. World Health Organization declares global emergency: a review of the 2019 novel coronavirus (COVID-19) Int J Surg. 2020;76:71–76. doi: 10.1016/j.ijsu.2020.02.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mirza MU, Froeyen M. Structural elucidation of SARS-CoV-2 vital proteins: computational methods reveal potential drug candidates against main protease, Nsp12 polymerase and Nsp13 helicase. J Pharm Anal. 2020 doi: 10.1016/j.jpha.2020.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ye Q, Wang B, Zhang T, Xu J, Shang S. The mechanism and treatment of gastrointestinal symptoms in patients with COVID-19. Am J Physiol Gastrointest Liver Physiol. 2020;319(2):G245–G252. doi: 10.1152/ajpgi.00148.2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mondal R, Lahiri D, Deb S, Bandyopadhyay D, Shome G, Sarkar S, Paria SR, Thakurta TG, Singla P, Biswas SC. COVID-19: are we dealing with a multisystem vasculopathy in disguise of a viral infection? J Thromb Thrombolysis. 2020 doi: 10.1007/s11239-020-02210-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lax SF, Skok K, Zechner P, Kessler HH, Kaufmann N, Koelblinger C, Vander K, Bargfrieder U, Trauner M. Pulmonary arterial thrombosis in COVID-19 with fatal outcome: results from a prospective, single-center, clinicopathologic case series. Ann Intern Med. 2020 doi: 10.7326/M20-2566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Organization WH. Modes of transmission of virus causing COVID-19: implications for IPC precaution recommendations: scientific brief, 27 March 2020. World Health Organization; 2020. [Google Scholar]
- 9.int/emergencies/diseases/novel-coronavirus-/situation-reports WHOJUhww Coronavirus disease (COVID-2019) situation reports. 2020.
- 10.Wang KY, Liu F, Jiang R, Yang X, You T, Liu X, Xiao CQ, Shi Z, Jiang H, Rao Z, Yang H (2020) Structure of Mpro from COVID-19 virus and discovery of its inhibitors. Nature. 582(7811): 289-93. [DOI] [PubMed]
- 11.Mukherjee PK, Bahadur S, Harwansh RK, Biswas S, Banerjee SJPR. Paradigm shift in natural product research: traditional medicine inspired approaches. 2017;16(5):803–826. [Google Scholar]
- 12.Wyganowska-Swiatkowska M, Nohawica M, Grocholewicz K, Nowak G Influence of herbal medicines on HMGB1 release, SARS-CoV-2 viral attachment, acute respiratory failure, and sepsis. Lit Rev. 2020;21(13):4639. doi: 10.3390/ijms21134639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Enmozhi SK, Raja K, Sebastine I, Joseph J (2020) Andrographolide as a potential inhibitor of SARS-CoV-2 main protease: an in silico approach. J Biomol Struct Dyn 1–7 [DOI] [PMC free article] [PubMed]
- 14.Suryanarayana L, Banavath D (2020) A review on identification of antiviral potential medicinal plant compounds against with COVID-19. Int J Res Eng Sci Manag 3(3):675–679
- 15.Kapepula PM, Kabengele JK, Kingombe M, Van Bambeke F, Tulkens PM, Sadiki Kishabongo A, Decloedt E, Zumla A, Tiberi S, Suleman F, Tshilolo L (2020) Artemisia spp. derivatives for COVID-19 treatment: anecdotal use, political hype, treatment potential, challenges, and road map to randomized clinical trials. Am J Trop Med Hyg 200820 [DOI] [PMC free article] [PubMed]
- 16.Samuels A (2020) Coronavirus Act 2020: An overview by a lawyer interested in medico-legal matters. Medico-Legal Journal. 88(2): 86-9. [DOI] [PubMed]
- 17.Joshi RS, Jagdale SS, Bansode SB, Shankar SS, Tellis MB, Pandya VK, Chugh A, Giri AP, Kulkarni MJ (2020) Discovery of potential multi-target-directed ligands by targeting host-specific SARS-CoV-2 structurally conserved main protease. J Biomol Struct Dyn 39(9): 3099-3144. [DOI] [PMC free article] [PubMed]
- 18.Haggag YA, El-Ashmawy NE, Okasha KM (2020) Is hesperidin essential for prophylaxis and treatment of COVID-19 Infection? Med Hypotheses 109957 [DOI] [PMC free article] [PubMed]
- 19.Alagu Lakshmi S, Shafreen RMB, Priya A, Shunmugiah KP (2020) Ethnomedicines of Indian origin for combating COVID-19 infection by hampering the viral replication: using structure-based drug discovery approach. J Biomol Struct Dyn 1–16 [DOI] [PMC free article] [PubMed]
- 20.Akamatsu H, Komura J, Asada Y, Niwa Y (1991) Mechanism of anti-inflammatory action of glycyrrhizin: effect on neutrophil functions including reactive oxygen species generation. Plant Med 57(02):119–121 [DOI] [PubMed]
- 21.Li W, Ashok M, Li J, Yang H, Sama AE, Wang H. A major ingredient of green tea rescues mice from lethal sepsis partly by inhibiting HMGB1. PLoS One. 2007;2(11):e1153. doi: 10.1371/journal.pone.0001153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yu B, Dai CQ, Jiang ZY, Li EQ, Chen C, Wu XL, Chen J, Liu Q, Zhao CL, He JX, Ju DH, Chen XY. Andrographolide as an anti-H1N1 drug and the mechanism related to retinoic acid-inducible gene-I-like receptors signaling pathway. Chin J Integr Med. 2014;20(7):540–545. doi: 10.1007/s11655-014-1860-0. [DOI] [PubMed] [Google Scholar]
- 23.Liu YT, Chen HW, Lii CK, Jhuang JH, Huang CS, Li ML, Yao HT (2020) A Diterpenoid, 14-Deoxy-11, 12-didehydroandrographolide, in andrographis paniculata reduces steatohepatitis and liver injury in mice fed a high-fat and high-cholesterol diet. Nutrients 12(2). 10.3390/nu12020523 [DOI] [PMC free article] [PubMed]
- 24.Liu Z, Xiao X, Wei X, Li J, Yang J, Tan H, Zhu J, Zhang Q, Wu J, Liu L. Composition and divergence of coronavirus spike proteins and host ACE2 receptors predict potential intermediate hosts of SARS-CoV-2. J Med Virol. 2020;92(6):595–601. doi: 10.1002/jmv.25726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li SY, Chen C, Zhang HQ, Guo HY, Wang H, Wang L, Zhang X, Hua SN, Yu J, Xiao PG, Li RS, Tan X. Identification of natural compounds with antiviral activities against SARS-associated coronavirus. Antiviral Res. 2005;67(1):18–23. doi: 10.1016/j.antiviral.2005.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wen CC, Kuo YH, Jan JT, Liang PH, Wang SY, Liu HG, Lee CK, Chang ST, Kuo CJ, Lee SS, Hou CC, Hsiao PW, Chien SC, Shyur LF, Yang NS. Specific plant terpenoids and lignoids possess potent antiviral activities against severe acute respiratory syndrome coronavirus. J Med Chem. 2007;50(17):4087–4095. doi: 10.1021/jm070295s. [DOI] [PubMed] [Google Scholar]
- 27.Cinatl J, Morgenstern B, Bauer G, Chandra P, Rabenau H, Doerr HW. Glycyrrhizin, an active component of liquorice roots, and replication of SARS-associated coronavirus. Lancet. 2003;361(9374):2045–2046. doi: 10.1016/s0140-6736(03)13615-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fiore C, Eisenhut M, Krausse R, Ragazzi E, Pellati D, Armanini D, Bielenberg J. Antiviral effects of Glycyrrhiza species. Phytother Res. 2008;22(2):141–148. doi: 10.1002/ptr.2295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yang MW, Chen F, Zhu DJ, Li JZ, Zhu JL, Zeng W, Qu SL, Zhang Y. Clinical efficacy of matrine and sodium chloride injection in treatment of 40 cases of COVID-19. Zhongguo Zhong Yao Za Zhi. 2020;45(10):2221–2231. doi: 10.19540/j.cnki.cjcmm.20200323.501. [DOI] [PubMed] [Google Scholar]
- 30.Kim DE, Min JS, Jang MS, Lee JY, Shin YS, Song JH, Kim HR, Kim S, Jin YH, Kwon S (2019) Natural bis-benzylisoquinoline alkaloids-tetrandrine, fangchinoline, and cepharanthine, inhibit human coronavirus OC43 infection of MRC-5 human lung cells. Biomolecules 9(11). 10.3390/biom9110696 [DOI] [PMC free article] [PubMed]
- 31.Ryu YB, Jeong HJ, Kim JH, Kim YM, Park JY, Kim D, Nguyen TT, Park SJ, Chang JS, Park KH, Rho MC, Lee WS. Biflavonoids from Torreya nucifera displaying SARS-CoV 3CL(pro) inhibition. Bioorg Med Chem. 2010;18(22):7940–7947. doi: 10.1016/j.bmc.2010.09.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yan H, Ma L, Wang H, Wu S, Huang H, Gu Z, Jiang J, Li Y. Luteolin decreases the yield of influenza A virus in vitro by interfering with the coat protein I complex expression. J Nat Med. 2019;73(3):487–496. doi: 10.1007/s11418-019-01287-7. [DOI] [PubMed] [Google Scholar]
- 33.Harwansh RK, Bahadur S. Herbal medicines to fight against COVID-19: new battle with an old weapon. Curr Pharm Biotechnol. 2022;23(2):235–260. doi: 10.2174/1389201022666210322124348. [DOI] [PubMed] [Google Scholar]
- 34.Wickramasinghe ASD, Kalansuriya P, Attanayake AP. Herbal medicines targeting the improved beta-cell functions and beta-cell regeneration for the management of diabetes mellitus. Evid Based Complement Alternat Med. 2021;2021:2920530. doi: 10.1155/2021/2920530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Agrawal N, Goyal A. Potential candidates against COVID-19 targeting RNA-dependent RNA polymerase: a comprehensive review. Curr Pharm Biotechnol. 2022;23(3):396–419. doi: 10.2174/1389201022666210421102513. [DOI] [PubMed] [Google Scholar]
- 36.Joshi T, Sharma P, Joshi T, Pundir H, Mathpal S, Chandra S. Structure-based screening of novel lichen compounds against SARS Coronavirus main protease (Mpro) as potentials inhibitors of COVID-19. Mol Divers. 2021;25(3):1665–1677. doi: 10.1007/s11030-020-10118-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Attah AF, Fagbemi AA, Olubiyi O, Dada-Adegbola H, Oluwadotun A, Elujoba A, Babalola CP. Therapeutic potentials of antiviral plants used in traditional african medicine with COVID-19 in focus: a Nigerian perspective. Front Pharmacol. 2021;12:596855. doi: 10.3389/fphar.2021.596855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Joshi C, Chaudhari A, Joshi C, Joshi M, Bagatharia S (2021) Repurposing of the herbal formulations: molecular docking and molecular dynamics simulation studies to validate the efficacy of phytocompounds against SARS-CoV-2 proteins. J Biomol Struct Dyn 1–15. 10.1080/07391102.2021.1922095 [DOI] [PMC free article] [PubMed]
- 39.Berman HM, Westbrook J, Feng Z, Gilliland G, Bhat TN, Weissig H, Shindyalov IN, Bourne PE. The Protein Data Bank. Nucleic Acids Res. 2000;28(1):235–242. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wu C, Liu Y, Yang Y, Zhang P, Zhong W, Wang Y, Wang Q, Xu Y, Li M, Li X, Zheng M, Chen L, Li H. Analysis of therapeutic targets for SARS-CoV-2 and discovery of potential drugs by computational methods. Acta Pharm Sin B. 2020 doi: 10.1016/j.apsb.2020.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mujwar S, Pardasani KR (2015) Prediction of riboswitch as a potential drug target and design of its optimal inhibitors for Mycobacterium tuberculosis. Int J Comput Biol Drug Des 8(4):326–347
- 42.Mujwar S, Pardasani KR (2015) Prediction of Riboswitch as a potential drug target for infectious diseases: an in silico case study of anthrax. J Med Imaging Health Inf 5(1):7–16
- 43.Shah K, Mujwar S, Gupta JK, Shrivastava SK, Mishra P. Molecular docking and in silico cogitation validate mefenamic acid prodrugs as human cyclooxygenase-2 inhibitor. Assay Drug Dev Technol. 2019;17(6):285–291. doi: 10.1089/adt.2019.943. [DOI] [PubMed] [Google Scholar]
- 44.Mujwar S, Deshmukh R, Harwansh RK, Gupta JK, Gour A. Drug repurposing approach for developing novel therapy against mupirocin-resistant Staphylococcus aureus. Assay Drug Dev Technol. 2019;17(7):298–309. doi: 10.1089/adt.2019.944. [DOI] [PubMed] [Google Scholar]
- 45.Minaz N, Razdan R, Hammock BD, Mujwar S, Goswami SK. Impact of diabetes on male sexual function in streptozotocin-induced diabetic rats: protective role of soluble epoxide hydrolase inhibitor. Biomed Pharmacother. 2019;115:108897. doi: 10.1016/j.biopha.2019.108897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mujwar S. Computational bioprospecting of andrographolide derivatives as potent cyclooxygenase-2 inhibitors. Biomed Biotechnol Res J. 2021;5(4):446. doi: 10.4103/bbrj.bbrj_56_21. [DOI] [Google Scholar]
- 47.Shah K, Mujwar S, Krishna G, Gupta JK. Computational design and biological depiction of novel naproxen derivative. Assay Drug Dev Technol. 2020;18(7):308–317. doi: 10.1089/adt.2020.977. [DOI] [PubMed] [Google Scholar]
- 48.Mujwar S, Shah K, Gupta JK, Gour A. Docking based screening of curcumin derivatives: a novel approach in the inhibition of tubercular DHFR. Int J Comput Biol Drug Des. 2021;14(4):297–314. doi: 10.1504/IJCBDD.2021.118830. [DOI] [Google Scholar]
- 49.Mujwar S, Kumar V. Computational drug repurposing approach to identify potential fatty acid-binding protein-4 inhibitors to develop novel antiobesity therapy. Assay Drug Dev Technol. 2020;18(7):318–327. doi: 10.1089/adt.2020.976. [DOI] [PubMed] [Google Scholar]
- 50.Mujwar S, Tripathi A (2021) Repurposing Benzbromarone as antifolate to develop novel antifungal therapy for Candida albicans. [DOI] [PMC free article] [PubMed]
- 51.Pires DE, Blundell TL, Ascher DB. pkCSM: predicting small-molecule pharmacokinetic and toxicity properties using graph-based signatures. J Med Chem. 2015;58(9):4066–4072. doi: 10.1021/acs.jmedchem.5b00104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Benet LZ, Hosey CM, Ursu O, Oprea TI. BDDCS, the rule of 5 and drugability. Adv Drug Deliv Rev. 2016;101:89–98. doi: 10.1016/j.addr.2016.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Veber DF, Johnson SR, Cheng HY, Smith BR, Ward KW, Kopple KD. Molecular properties that influence the oral bioavailability of drug candidates. J Med Chem. 2002;45(12):2615–2623. doi: 10.1021/jm020017n. [DOI] [PubMed] [Google Scholar]
- 54.System SR-DMD (2020) Maestro-Desmond interoperability tools, Schrödinger, New York, NY, 2021. D. E. Shaw Research, New York, NY, 2021
- 55.Bowers KJ, Chow DE, Xu H, Dror RO, Eastwood MP, Gregersen BA, Klepeis JL, Kolossvary I, Moraes MA, Sacerdoti FD, Salmon JK (2006) Scalable algorithms for molecular dynamics simulations on commodity clusters. In SC'06: Proceedings of the 2006 ACM/IEEE Conference on Supercomputing, IEEE, pp 43–43
- 56.Roos K, Wu C, Damm W, Reboul M, Stevenson JM, Lu C, Dahlgren MK, Mondal S, Chen W, Wang L, Abel R, Friesner RA, Harder ED. OPLS3e: extending force field coverage for drug-like small molecules. J Chem Theory Comput. 2019;15(3):1863–1874. doi: 10.1021/acs.jctc.8b01026. [DOI] [PubMed] [Google Scholar]
- 57.Berendsen H, Grigera J, Straatsma TP (1987) The missing term in effective pair potentials. J Phys Chem 91(24):6269–6271
- 58.Toukan K, Rahman A. Molecular-dynamics study of atomic motions in water. Phys Rev B Condens Matter. 1985;31(5):2643–2648. doi: 10.1103/physrevb.31.2643. [DOI] [PubMed] [Google Scholar]
- 59.Gahtori J, Pant S, Srivastava HK. Modeling antimalarial and antihuman African trypanosomiasis compounds: a ligand- and structure-based approaches. Mol Divers. 2020;24(4):1107–1124. doi: 10.1007/s11030-019-10015-y. [DOI] [PubMed] [Google Scholar]
- 60.Posch HA, Hoover WG, Vesely FJ. Canonical dynamics of the Nose oscillator: stability, order, and chaos. Phys Rev A Gen Phys. 1986;33(6):4253–4265. doi: 10.1103/physreva.33.4253. [DOI] [PubMed] [Google Scholar]
- 61.Martyna GJ, Tobias DJ, Klein ML (1994) Constant pressure molecular dynamics algorithms. J Chem Phys 101(5):4177–4189
- 62.Petersen HG (1995) Accuracy and efficiency of the particle mesh Ewald method. J Chem Phys 103(9):3668–3679
- 63.Mujwar S. Computational repurposing of tamibarotene against triple mutant variant of SARS-CoV-2. Comput Biol Med. 2021;136:104748. doi: 10.1016/j.compbiomed.2021.104748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Pradhan P, Soni NK, Chaudhary L, Mujwar S, Pardasani KR. In-silico prediction of riboswitches and design of their potent inhibitors for H1N1, H2N2 and H3N2 strains of influenza virus. Biosci, Biotechnol Res Asia. 2015;12(3):2173–2186. doi: 10.13005/bbra/1889. [DOI] [Google Scholar]
- 65.Soni N, Pardasani KR, Mujwar S. In silico analysis of dietary agents as anticancer inhibitors of insulin like growth factor 1 receptor (IGF1R) J Pharm Pharm Sci. 2015;7(9):191–196. [Google Scholar]
- 66.Jain R, Mujwar S. Repurposing metocurine as main protease inhibitor to develop novel antiviral therapy for COVID-19. J Structural Chemistry. 2020;31(6):2487–2499. doi: 10.1007/s11224-020-01605-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Woo CSJ, Lau JSH, El-Nezami H (2012) Herbal medicine: toxicity and recent trends in assessing their potential toxic effects. In Advances in Botanical Research, vol 62. Elsevier, pp 365–384
- 68.Kar P, Sharma NR, Singh B, Sen A, Roy A (2021) Natural compounds from Clerodendrum spp. as possible therapeutic candidates against SARS-CoV-2: an in silico investigation. J Biomol Struct Dyn 39(13):4774–4785 [DOI] [PMC free article] [PubMed]
- 69.Kar P, Sharma NR, Singh B, Sen A, Roy A (2020) Natural compounds from Clerodendrum spp. as possible therapeutic candidates against SARS-CoV-2: an in silico investigation. J Biomol Struct Dyn1–12 [DOI] [PMC free article] [PubMed]
- 70.Wang Y, Bryant SH, Cheng T, Wang J, Gindulyte A, Shoemaker BA, Thiessen PA, He S, Zhang J. PubChem BioAssay: 2017 update. Nucleic Acids Res. 2017;45(D1):D955–D963. doi: 10.1093/nar/gkw1118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Murugan NA, Pandian CJ, Jeyakanthan J (2020) Computational investigation on Andrographis paniculata phytochemicals to evaluate their potency against SARS-CoV-2 in comparison to known antiviral compounds in drug trials. J Biomol Struct Dyn 1–12 [DOI] [PMC free article] [PubMed]
- 72.Khaerunnisa S, Kurniawan H, Awaluddin R, Suhartati S, Soetjipto S (2020) Potential inhibitor of COVID-19 main protease (Mpro) from several medicinal plant compounds by molecular docking study. Preprints 20944:1–14
- 73.Kumar N, Singh A, Gulati HK, Bhagat K, Kaur K, Kaur J, Dudhal S, Duggal A, Gulati P, Singh H, Singh VJ (2021) Phytoconstituents from ten natural herbs as potent inhibitors of main protease enzyme of SARS-COV-2: in silico study. Phytomed Plus 1(4):100083 [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Yes.