Figure 4. Determination of CDC assay parameters.
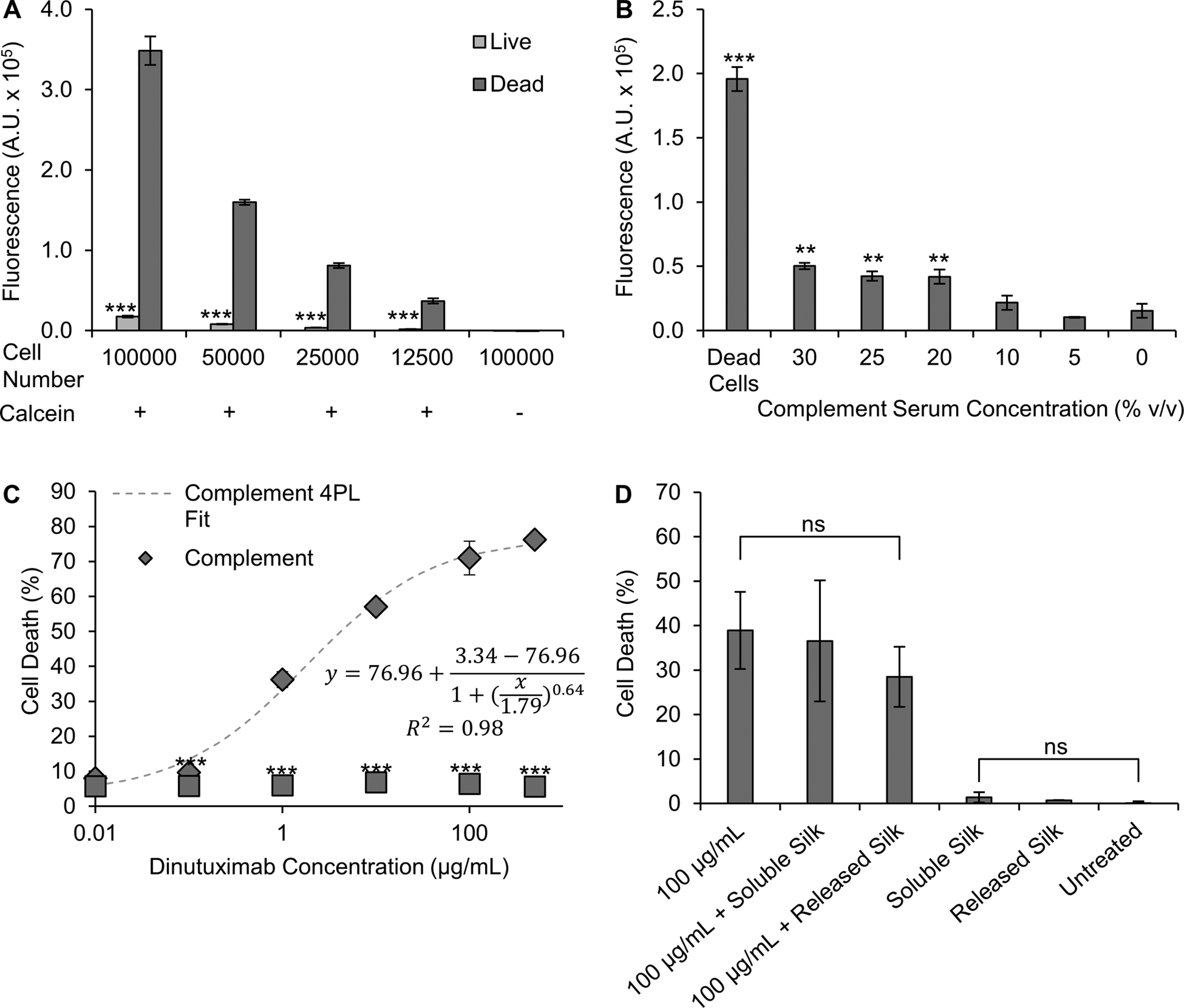
Identification of (A) number of cells needed for CDC assay, (B) toxicity of complement serum, and (C) toxicity of DNX with and without complement serum. (D) Interference of silk formats with the assay was investigated via soluble silk added to the culture medium (silk) or supernatant from silk foam release experiments (silk release; note these are control samples not loaded with DNX). The presence of silk did not interfere with the assay. Data are presented as mean ± standard deviation of three independent samples. (A) *** p<0.001 as compared to dead cells based on a t-test. (B) ** p<0.01, *** p<0.001 when compared to non-complement serum treatment based on one‐way ANOVA followed by Tukey’s post hoc test. (C) *** p<0.001 as compared to DNX-matched concentration with complement serum based on a t-test.
