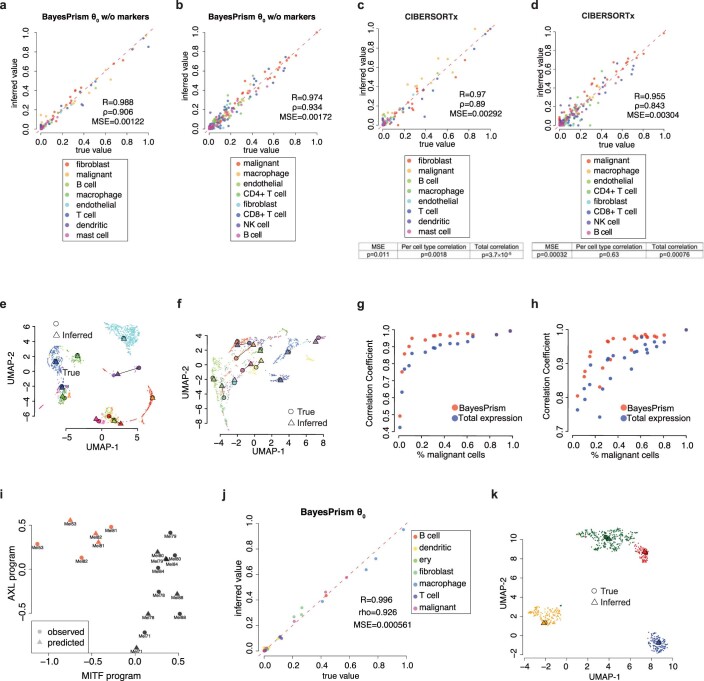
Extended Data Fig. 4. Performance of BayesPrism in inferring cell type composition and gene expression in malignant cells on the leave-one-out pseudo-bulk data of HNSCC, SKCM and OV.
(a-b) Scatter plots show θ0 in (a) HNSCC and (b) SKCM versus the ground truth in pseudo-bulk. (c-d) Scatter plots show the CIBERSORTx inferred cell type fraction in (c) HNSCC and (d) SKCM versus the ground truth in pseudo-bulk. Tables below show the one-sided p values for cell type fractions inferred by BayesPrism and CIBERSORTx. T test was used for MSE and z test was performed on Fisher’s Z-transformed cell type-level correlation coefficients (see Methods). (e-f) UMAP shows the expression profile of individual malignant cells in scRNA-seq of (e) HNSCC and (f) SKCM colored by patient ID. Patients with >50 malignant cells and cells with reads detected for >3000 genes are shown. The inferred expression profile, shown as △, and the averaged expression profile from scRNA-seq for each patient, shown as ○, are projected onto the UMAP manifold. (g-h) Scatter plot shows Pearson’s correlation coefficient between inferred expression and that of the averaged expression from malignant cells in scRNA-seq of (g) HNSCC and (h) SKCM as a function of the fraction of malignant cells in each simulated data. The correlation coefficient was computed on DESeq2 variance-stabilized transformed values. Red marks the correlation inferred by BayesPrism, while blue marks that of total expression of the simulated data. (i) Scatter plot shows the AXL and MITF program scores of malignant cells from SKCM in the leave-one-out test, calculated by the mean of Z scores over the corresponding marker genes. ○ marks the average expression of malignant cells in each patient from the scRNA-seq data, while △ marks the inferred expression profile of malignant cells in each patient. (j) Scatter plots show θ0 in OV versus the ground truth in pseudo-bulk. (k) UMAP shows the expression profile of individual malignant cells in scRNA-seq of OV colored by patient ID. The inferred expression profile, shown as △, and the averaged expression profile from scRNA-seq for each patient, shown as ○, are projected onto the UMAP manifold.