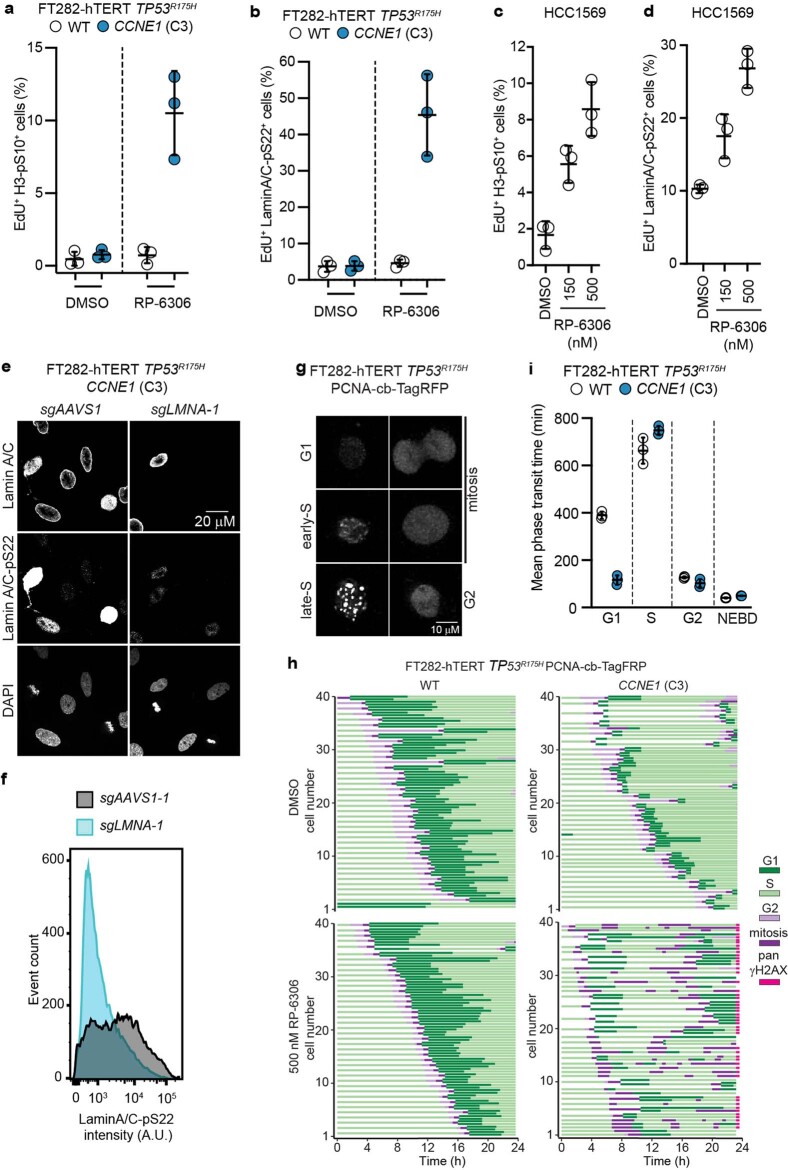
Extended Data Fig. 5. RP-6306 causes unscheduled mitosis in CCNE1-high cells.
a,b, Quantitation of double-positive staining for EdU and either histone H3-pS10 (a) or Lamin A/C-pS22 (b) by FACS following vehicle (DMSO) or RP-6306 treatment (500 nM) for 24 h. Data are shown as mean ± s.d. (n = 3). The gating strategy is shown in Supplementary Figure 2. c,d, Quantitation of double-positive staining for EdU and either histone H3-pS10 (c) or Lamin A/C-pS22 (d) by FACS in HCC1569 cells following vehicle (DMSO) or the indicated dose of RP-6306 for 24 h. Data are shown as mean ± s.d. (n = 3). The gating strategy is shown in Extended Data Fig. 6b. e,f, Validation of the lamin A/C-pS22 antibody. (e) Representative micrographs of FT282-hTERT TP53R175H CCNE1 cells transduced with lentivirus expressing sgRNAs targeting either AAVS1 (sgAAVS1) or LMNA (sgLMNA-1) stained with DAPI and the indicated antibodies. (f) Flow cytometry histograms of the same cells showing loss of Lamin A/C-pS22 signal in the sgLMNA-1 condition. g–i, Time-lapse imaging of FT282-hTERT TP53R175H PCNA-chromobody-TagRFP (WT) and CCNE1-high (CCNE1) cells treated with DMSO or RP-6306 (500 nM) for 23 h. (g) Pattern of PCNA localization used to identify cell cycle stages. (h) Cell cycle profile of individual cells (each bar represents one cell). 30 cells were analysed at random for each condition. (i) Quantitation of cell cycle phase transit time of indicated samples treated with DMSO for 23 h. Data are shown as mean ± s.d. (n = 3).