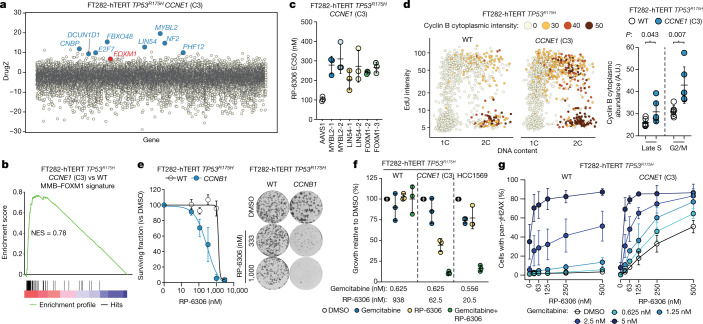
Fig. 4. Replication stress and FOXM1–MMB activity underlie vulnerability to PKMYT1 inhibition.
a, RP-6306 resistance screen for dose required to kill 80% of cells (LD80) performed in FT282-hTERT TP53R175H CCNE1 (C3 and C4) with DrugZ scores for C3 plotted. Genes with DrugZ > 9 in both C3 and C4 screens (blue) and FOXM1 (red) are shown for reference. b, Gene set enrichment analysis (GSEA) of differential gene expression in FT282 parental (WT) versus CCNE1-high (C3) cells for genes co-regulated by MMB–FOXM1. c, EC50 values for RP-6306 in CCNE1-high FT282-hTERT TP53R175H cells nucleofected with Cas9 ribonucleoproteins assembled with the indicated sgRNAs. Growth was monitored by clonogenic survival assay. d, QIBC analysis of cyclin B cytoplasmic intensity, EdU incorporation and DNA content (measured with DAPI). Representative QIBC plots (left) and cytoplasmic cyclin B intensity (right) quantification in late S or G2/M. P values determined by two-tailed t-test. e, Clonogenic survival of FT282-hTERT TP53R175H CCNB1-2A-GFP and wild-type parental cells treated with RP-6306. Quantification (left) and representative images of plates stained with crystal violet (right). f, Growth inhibition relative to DMSO control of parental (WT) and CCNE1-high FT282-hTERT TP53R175H cells and HCC1569 cells after the indicated treatments. Growth was monitored with an Incucyte live-cell imager for up to six population doublings. g, QIBC quantification of cells with pan-γH2AX in response to the indicated RP-6306–gemcitabine combinations. Data in c–g are mean ± s.d. (n = 3).