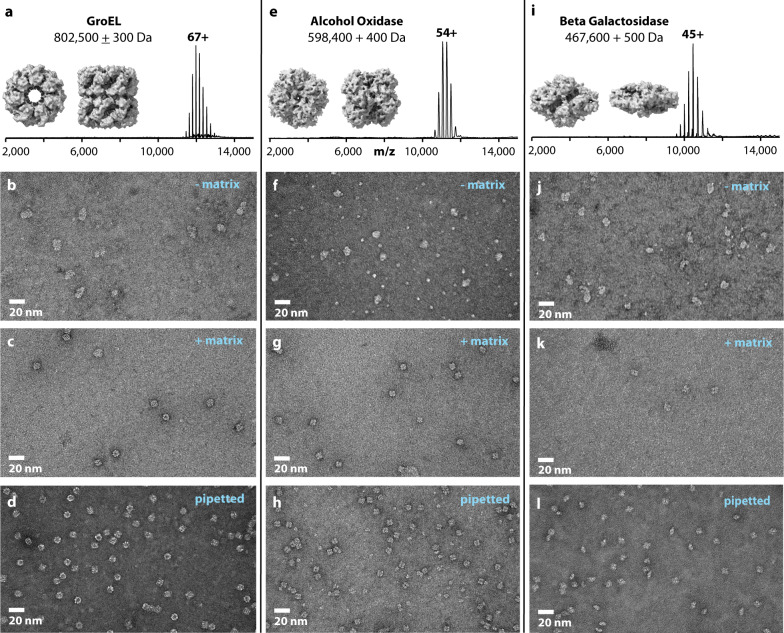
Fig. 1. Matrix-landed protein complexes retain structural features and integrity as imaged by negative stain TEM.
a Native mass spectrum of GroEL complexes along with molecular model images of GroEL. b Negative stain TEM image of GroEL ions landed onto bare carbon TEM grids. c Negative stain TEM image of GroEL ions matrix-landed onto TEM grids having a thin film of glycerol. d Negative stain TEM image of GroEL molecules that were conventionally prepared. e Native mass spectrum of alcohol oxidase complexes along with molecular model images of alcohol oxidase. f, g Negative stain TEM images of alcohol oxidase ions landed on bare carbon TEM grids (f), or matrix-landing TEM grids coated with a thin film of glycerol (g). h Conventionally prepared negative stain TEM images of alcohol oxidase molecules. i Native mass spectrum the b-galactosidase complex along with molecular model images of b-galactosidase. j, k Negative stain TEM images of b-galactosidase ions landed onto either bare carbon TEM grids (j) or matrix-landing TEM grids coated with a thin film of glycerol (k). l Conventionally prepared negative stain TEM images of b-galactosidase molecules. Note the structural models contained in panels (a, e, and I) were generated from PDB structures (PDB:5W0S, PDB:6H3G, PDB:6X1Q) and included here to aid in image interpretation21, 26, 27. Images similar to those shown in panels (b–l) were acquired no fewer than five times each.