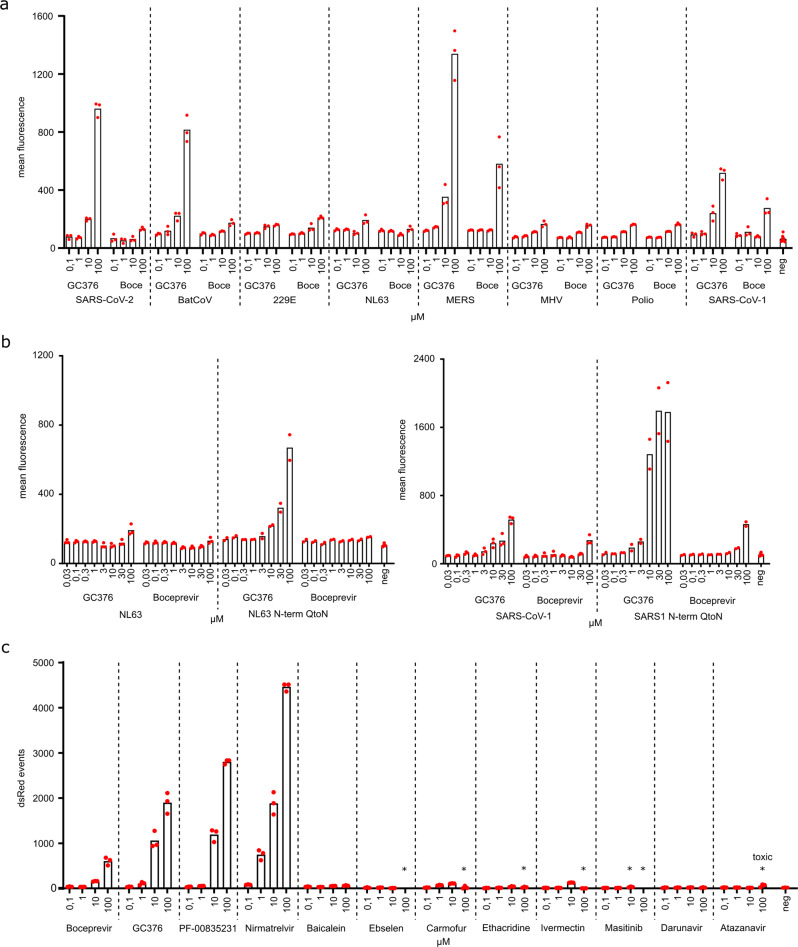
Fig. 4. Inhibition-on assay is adaptable to further proteases and helps dissect potent and proposed inhibitors.
a Seven proteases from close and distantly related viruses to SARS-CoV-2 (Rousettus bat coronavirus HKU9 (BatCoV), human coronaviruses 229E and NL63, MERS, mouse hepatitis virus (MHV), poliovirus, and SARS-CoV-1) were tested in 293T cells transfected with inhibition-on assay plasmids and treated with the compounds GC376 and boceprevir (abbreviated: Boce). Mean fluorescent intensity of positive cells was used to display read-outs (Supplementary Figs. 5 and 6). (n = 3 biologically independent replicates per condition with average values represented by histogram bars; negative control (neg) = average of transfected cells of each construct without inhibitor; dashed lines separate different constructs). b N-terminal cis-cleavage glutamine to asparagine mutants of the same seven proteases were tested for increased susceptibility to compounds. NL63 and SARS-CoV-1 showed increased response. (Wild-type: n = 3 biologically independent replicates per condition with average values represented by histogram bars; N-term-Q-to-N: n = 2 biologically independent replicates; negative control (neg) = transfected cells of each construct without inhibitor; dashed lines separate different constructs). c A panel of compounds was tested via FACS. Red fluorescent live singlet cell events were chosen to display read-outs (Supplementary Figs. 5 and 6). An asterisk above the compound concentrations (*) indicates visible toxicity (Supplementary Fig. 4). (n = 3 biologically independent replicates per condition with average values represented by histogram bars; negative control (neg) = infected cells without inhibitor; dashed lines separate different inhibitors).