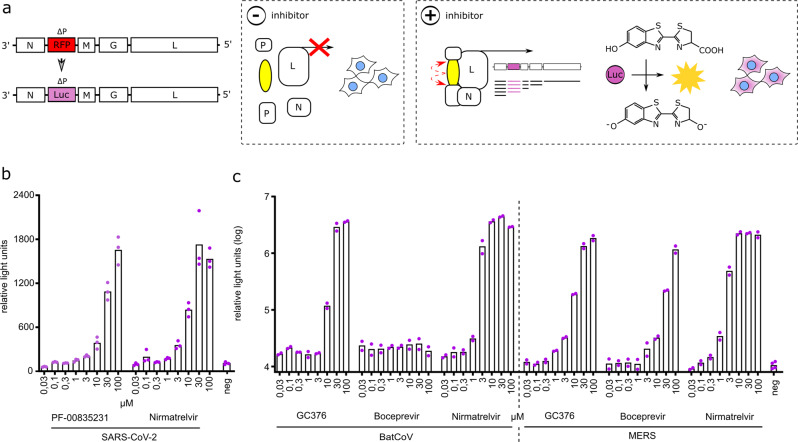
Fig. 5. Adaptation of inhibition-on assay to bioluminescence.
a The red fluorescent protein (RFP) dsRed in VSV-∆P was replaced with firefly luciferase, generating VSV-∆P-Luc. Similar to the previous inhibition-on assay, addition of an inhibitor facilitates viral replication and gene expression. (dashed boxes separate schematics of molecular mechanism occurring in untreated (− protease inhibitor) versus treated (+ protease inhibitor) cells). b 293T cells were transiently transfected with the SARS-CoV-2 inhibition-on plasmid and treated with either PF-00835231 or PF-07321332/nirmatrelvir and VSV-∆P-Luc. (n = 3 biologically independent replicates per condition with average values represented by histogram bars; negative control (neg) = average of transfected cells of each construct without inhibitor). c BHK21 stably transduced with bat coronavirus HKU9 (BatCoV) and MERS main proteases were tested with GC376, boceprevir, and PF-07321332/nirmatrelvir. (n = 2 biologically independent replicates per condition with average values represented by histogram bars; negative control (neg) = average of transfected cells of each construct without inhibitor; dashed lines separate differently transduced cells).