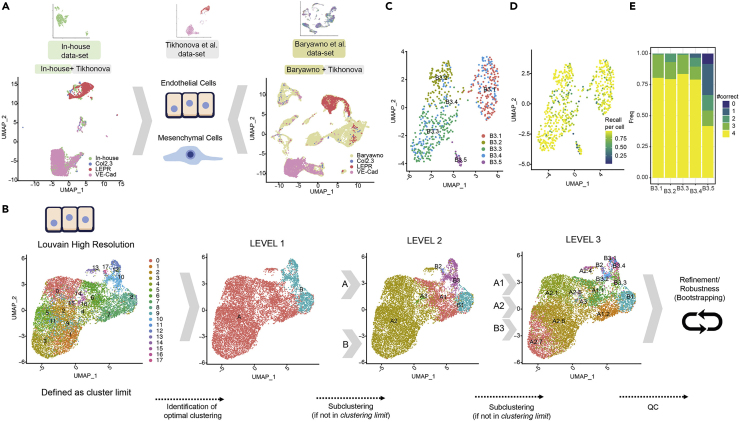
Figure 2.
Data integration and high-resolution clustering strategy
(A) Integrated analysis of the bone marrow niche datasets (two publicly available, Tikhonova et al., 2019 and Baryawno et al., 2019 and one generated in our lab, in-house) separately for two well-defined population (endothelial and mesenchymal cells). Tikhonova et al., 2019 dataset is used as a reference considering their separated cell profiling strategy for COL2.3+, LEPR+, and VE-Cad+ populations. In the top row, a UMAP projection is depicted for each single-cell RNA dataset. In the left-lower (“In-house + Tikhonova”) and in the right-lower (“Baryawno + Tikhonova”), datasets are integrated to identify endothelial and mesenchymal populations.
(B) Clustering strategy: the analysis of endothelial cells as an example. An upper limit to the cluster is set for the clustering (left panel) using Louvain high-resolution clustering. Then, an iterative divide-and-conquer strategy identifies the optimal level of clusters at different levels: Level 1 (second panel from the left), Level 2 (third panel), and Level 3 (fourth panel).
(C–E) The robustness analysis for sub-clustering B3 (from Level 2 to Level 3). Specifically: (C) subclusters identified, (D) the fraction of assignments to its original cluster using a random-forest + bootstrapping strategy and (E) summary of the results (D) per cluster, #correct indicates the times a cluster is a dominant cluster (see STAR Methods) for a cell within it in all pairwise comparisons (see Figure S2 for the sub-clustering analysis of A1 and A2). See also Figures S2 and S3.