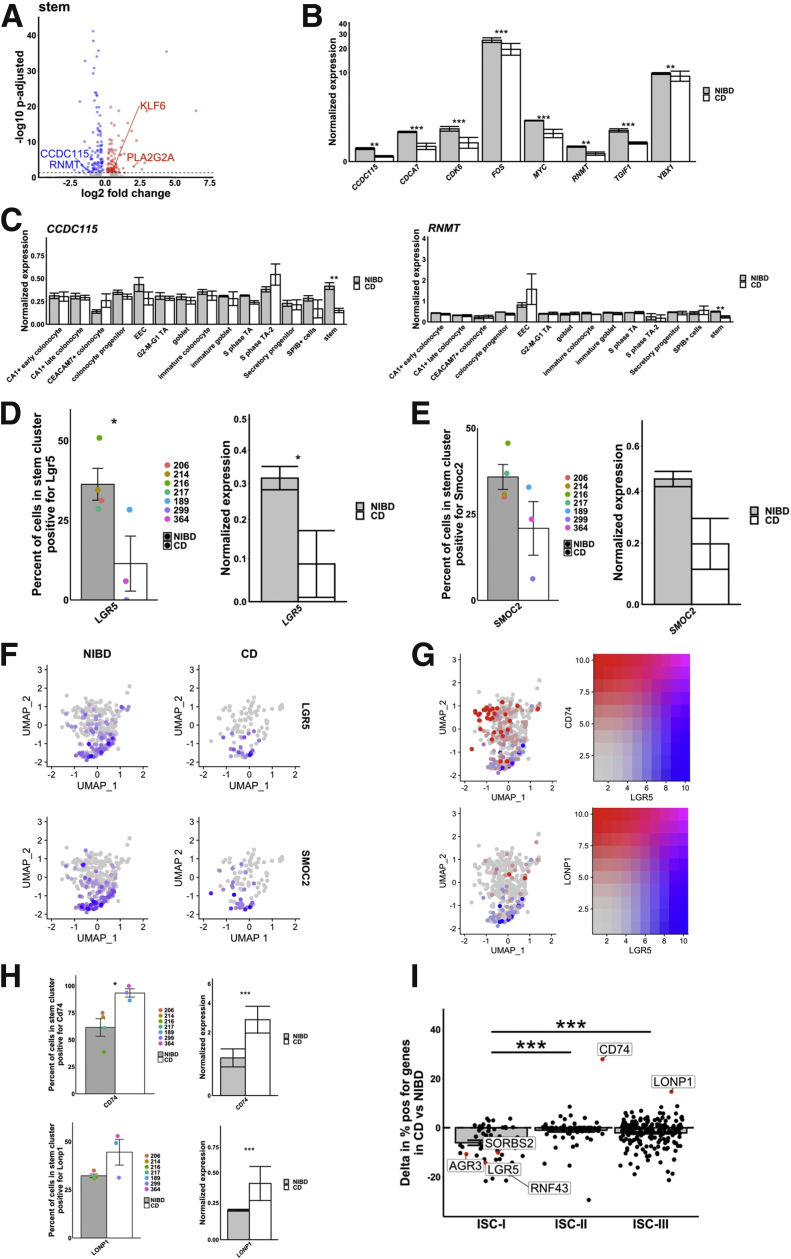
Figure 7.
CD disrupts stem cell homeostasis. (A) Differentially expressed gene within the stem cluster. Log2 fold change is shown on the x-axis and the adjusted P value is shown on the y-axis. (B) Mean expression of 8 mediators of the Wnt pathway in NIBD and CD samples show a consistent decrease in response to CD. (C) Mean expression of CCDC115 and RNMT across clusters shows a significant decrease only occurs in the stem cluster. Percentage of cells positive for (D) LGR5 (left) and (E) SMOC2 (left) or the average normalized expression for (D) LGR5 (right) and (E) SMOC2 (right) in the stem cluster for NIBD or CD samples. Dots relate to individual sample value. (F) Uniform Manifold Approximation and Projection (UMAP) of the stem cluster with expression of LGR5 or SMOC2 overlain, separated by NIBD/CD. (G) UMAP of the stem cluster with LGR5 expression (red) co-overlain with either CD74 (top) or LONP1 (bottom) (both blue) expression. Right: Scale for expression is shown. (H) Percentage of cells positive for CD74 (left top) and LONP1 (left bottom) or the average normalized expression for CD74 (right top) and LONP1 (right bottom) in the stem cluster for NIBD or CD samples. Dots relate to individual sample value. (I) Shift in percentage of positive genes in either ISC-I, ISC-II, or ISC-III cell types between NIBD and CD. P values for expression bar plots were calculated using the Wilcoxon rank-sum test and the P value for the ISC subtype shifts was calculated using a Student t test. ∗P < .05, ∗∗P < .01, and ∗∗∗P < .001.