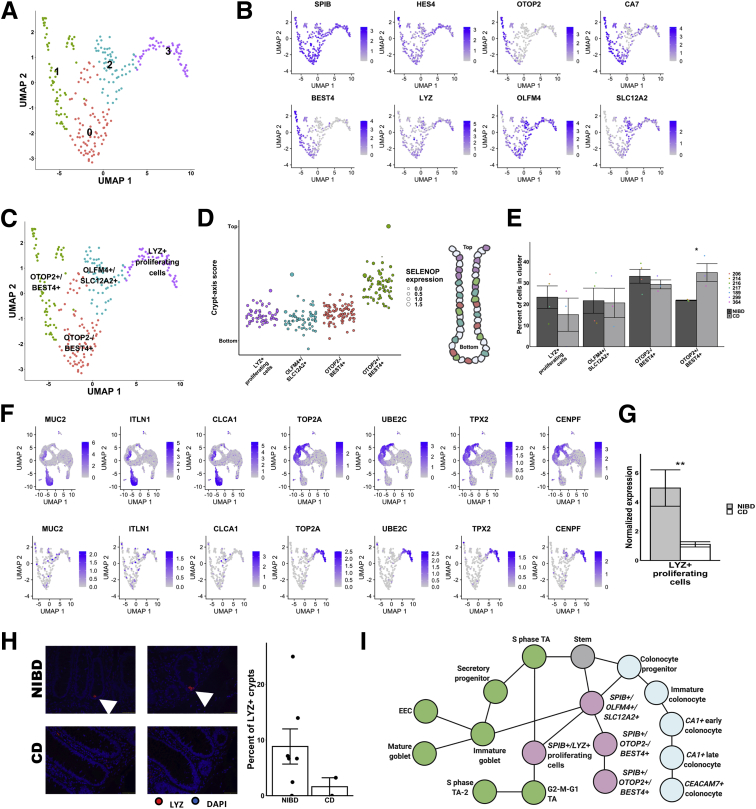
Figure 8.
Subcluster analysis of SPIB+ cells shows distinct lineages. (A) Uniform Manifold Approximation and Projection (UMAP) after subclustering of SPIB+ cells with labeling of the 4 identified subclusters. (B) UMAP of SPIB+ cell subclusters overlain with gene expression from genes marking the entire cluster or distinct subclusters. (C) SPIB+ cell subcluster UMAP following cell type assignment using highly enriched markers of the subclusters. (D) Crypt–axis scores (low near crypt bottom, high near crypt top) of subcluster-assigned cells uncovers a colonocyte signature in the OTOP2+/BEST4+ subcluster. Clusters are arranged on the x-axis by mean crypt–axis score. The size of the dot corresponds to the level of expression of SELENOP, a known marker of the top of the crypt. (E) Mean cluster cell abundances across NIBD and CD samples shows a significant increase in OTOP2+/BEST4+ subcluster in CD. Dots show abundances of individual samples. (F) UMAP of either all clusters (top) or SPIB+ cell subclusters (bottom) overlain with the expression of markers of secretory progenitor cells. (G) Mean normalized expression of LYZ in NIBD and CD samples in the LYZ+ proliferating subcluster. (H) LYZ-detected immunofluorescence in colonic crypts from either NIBD (n = 181 crypts, 5 patients) or CD (n = 76 crypts, 2 patients) patient examples (left) and quantified by sample (right). Mean percentage of crypts positive for LYZ in NIBD or CD. (I) Lineage reconstruction as determined by partition-based graph abstraction (PAGA). P values are calculated by a Student t test. ∗P < .05. DAPI, 4′,6-diamidino-2-phenylindole.