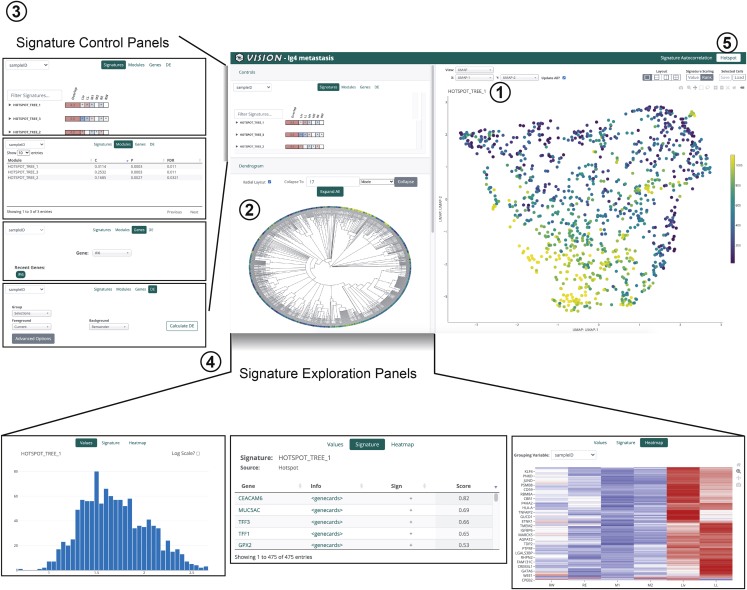
Figure 1.
Overview of the PhyloVision interactive UI
PhyloVision’s user interface (UI) is a web-based, feature-rich report that can be hosted locally or externally. PhyloVision incorporates four main panels into viewing. First is a panel for visualization of two-dimensional single-cell RNA-seq projections (e.g., a UMAP projection) or, alternatively, coordinates (e.g., from spatial transcriptomics datasets; inset 1). Second is a panel for interactive visualization of a phylogeny relating all cells (inset 2) that enables selection, collapsing, and variable layouts (radial or linear). Third is a control panel for selecting values to be overlaid onto the phylogeny and two-dimensional visualization panel, evaluating statistics associated with each signature or module, plotting a gene’s expression, or performing differential expression analysis, with each signature or module, plotting a gene’s expression, or performing differential expression analysis. In the default “Signature Autocorrelation” mode, signatures are clustered using a Gaussian mixture model to group together signatures with similar distributions (STAR Methods; inset 3). Note that here the Hotspot mode is shown, which operates “bottom up”: first finding heritable gene modules and then analyzing their over-representation (enrichment) in user-provided signatures. The alternative mode, “Signature Autocorrelation,” that operates directly on the user signatures is described in DeTomaso et al., 2019. Users can control the analysis mode by toggling between “Signature Autocorrelation” and “Hotspot” (inset 5). Fourth is an exploration panel for inspecting the value distribution, gene membership, and expression heatmaps of each user-provided gene signature or automatically identified Hotspot module (inset 4). See also Figures S1–S4 and Video S1.