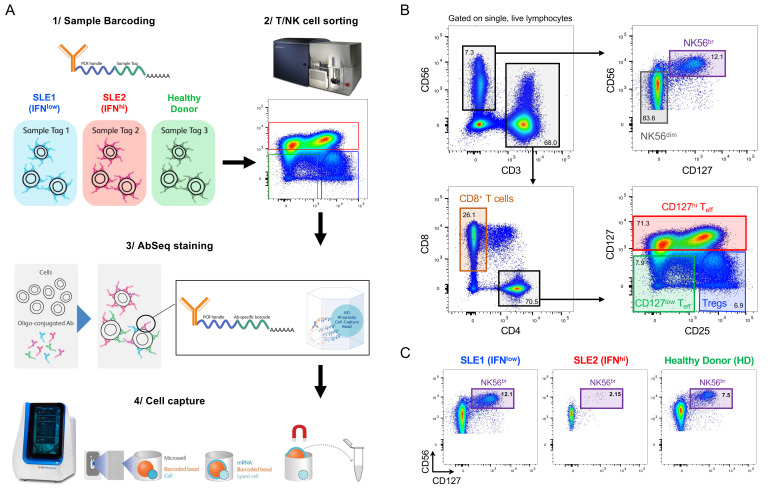
Figure 1. Characterisation of the peripheral T and NK cell populations in SLE using a single-cell multi-omics approach.
( A) Summary of the experimental workflow based on the BD Rhapsody single-cell RNA-sequencing system combining the quantification of mRNA and surface protein targets (AbSeq). Experiment was carried out using samples from three donors: (i) an SLE patient with low expression of the type I interferon (IFN)-induced gene expression signature (IFN low; depicted in blue); (ii) an SLE patient with a previously detected IFN-induced gene expression signature (IFN hi; depicted in red); and (iii) a healthy donor (HD; depicted in green). Cells isolated from each donor were barcoded with oligo-conjugated antibodies and pooled together for cell capture and library preparation. ( B) Gating strategy used for the isolation of the six T and NK cell populations assessed in this study. ( C) Frequency of NK56 br cells, defined as CD3 -CD127 +CD56 hi NK cells, in each of the three donors.