Abstract
Background
A rapid, accurate, non-invasive diagnostic screen is needed to identify people with SARS-CoV-2 infection. We investigated whether organic semi-conducting (OSC) sensors and trained dogs could distinguish between people infected with asymptomatic or mild symptoms, and uninfected individuals, and the impact of screening at ports-of-entry.
Methods
Odour samples were collected from adults, and SARS-CoV-2 infection status confirmed using RT-PCR. OSC sensors captured the volatile organic compound (VOC) profile of odour samples. Trained dogs were tested in a double-blind trial to determine their ability to detect differences in VOCs between infected and uninfected individuals, with sensitivity and specificity as the primary outcome. Mathematical modelling was used to investigate the impact of bio-detection dogs for screening.
Results
About, 3921 adults were enrolled in the study and odour samples collected from 1097 SARS-CoV-2 infected and 2031 uninfected individuals. OSC sensors were able to distinguish between SARS-CoV-2 infected individuals and uninfected, with sensitivity from 98% (95% CI 95–100) to 100% and specificity from 99% (95% CI 97–100) to 100%. Six dogs were able to distinguish between samples with sensitivity ranging from 82% (95% CI 76–87) to 94% (95% CI 89–98) and specificity ranging from 76% (95% CI 70–82) to 92% (95% CI 88–96). Mathematical modelling suggests that dog screening plus a confirmatory PCR test could detect up to 89% of SARS-CoV-2 infections, averting up to 2.2 times as much transmission compared to isolation of symptomatic individuals only.
Conclusions
People infected with SARS-CoV-2, with asymptomatic or mild symptoms, have a distinct odour that can be identified by sensors and trained dogs with a high degree of accuracy. Odour-based diagnostics using sensors and/or dogs may prove a rapid and effective tool for screening large numbers of people.
Trial Registration NCT04509713 (clinicaltrials.gov).
Keywords: COVID-19, rapid screening, public health, infection control
Introduction
To control the spread of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), which causes COVID-19, it is essential to rapidly and accurately screen infected individuals within communities, at ports-of-entry and other places where large numbers of people congregate and prevent onward transmission. As new variants emerge,1,2 the need for rapid and sensitive screening methods is increasingly important to ensure border biosecurity, instil confidence in people to travel and, re-opening and re-invigorate economies, which are dependent on travel or congregation of large numbers of people. Although quarantining plus testing by reverse transcription-polymerase chain reaction (RT-PCR) is done at some ports-of-entry, this strategy is slow, costly and cause inconvenience. Current testing methods for SARS-CoV-2 are unsuitable for rapid screening of large numbers of people, such as those found in airports and other public venues, due to low sensitivity and/or delays in the return of test results.3 Real-time RT-PCR testing remains the gold standard in SARS-CoV-2 diagnostics but is impractical for rapid screening due to long turn-around times. Real-time RT-PCR may also detect residual or degraded viral RNA past the point an individual is infectious.4 The Innova SARS-CoV-2 Antigen Rapid Qualitative Test, a lateral flow test (LFT), may detect individuals with viral loads high enough to cause onwards transmission (83% sensitivity)5 or those with culturable virus (98% sensitivity)6 however, may fail to detect lower viral loads.7 From a practical point of view, real-time RT-PCR and LFT testing may be unsuitable for mass screening since they are invasive, time consuming and costly.
There is evidence that infections with respiratory viruses produce distinct odour signatures that are pathogen specific and may be associated with an odour that could be used for disease detection.8–11 Using trained dogs or electronic sensors to identify people infected with SARS-CoV-2 by their odour, might prove to be a tractable method for mass screening passengers since they are non-invasive, able to screen people rapidly in real time,12 are low cost and potentially scalable. Bio-detection dogs could be used at places where rapid screening of a large number of people is required including ports-of-entry, train stations, sporting venues and places of work.12 Bio-detection dogs are increasingly deployed as an efficient, reliable, and mobile diagnostic tools to recognize volatile biomarkers contained in human breath, skin, and urine produced by specific diseases and chronic health conditions.13–16 Dogs have an extraordinary olfactory capability with odour detection 10 000–100 000 times higher than an average person, and the dog’s lower limit of detection of volatile organic compounds (VOCs) is one part per trillion.17 Several pilot studies suggest that dogs can detect hospitalised patients with SARS-CoV-2 in Colombia,18 France,19 Germany,20,21 Iran,22 Lebanon,19 United Arab Emirates23 and USA24 with a high sensitivity and/or specificity. Here, we aimed to assess whether there is a specific odour associated with infection with SARS-CoV-2, and whether trained dogs and organic semi-conducting (OSC) sensors can distinguish between the odour of uninfected individuals (real-time RT-PCR negative) and infected individuals (real-time RT-PCR positive) who are displaying mild symptoms or are asymptomatic (Appendix 2 pp 5, available as Supplementary data at JTM online). Modelling was also done to investigate the likely impact of dogs as part of a testing strategy.
Methods
Ethics approval
This study was done in accordance with the recommendations for physicians involved in research on human participants adopted by the 18th World Medical Assembly, Helsinki 1964 and later revisions. This study received full approval from the Health and Safety Executive (ref: CBA1.654.20.1, 16 June 2021), London School of Hygiene and Tropical Medicine Ethics Committee (ref: 22159, 22 June 2020) and Animal Welfare Ethics Review Board (ref: 2020-06 19-Jun-20), Department of Biosciences Ethics Committee, Durham University (19 June 2020), the Health Research Authority (ref: 284221, 17 June 2020), and relevant NHS trust Research Ethics Committees. The study was registered on clinicaltrials.gov (ID: NCT04509713).
Study design and participants
Participants were recruited from 2 July 2020 to 1 March 2021 in Great Britain using phone calls, text-messages, emails, leaflets/posters, videos, social media and press articles. The target sample size was 325 SARS-CoV-2 real-time RT-PCR positive (henceforth abbreviated as RT-PCR, infected) participants and at least 675 SARS-CoV-2 RT-PCR negative (uninfected) participants. National Health Service (NHS) staff and their household members were recruited through 23 NHS hospital sites and members of the public recruited via an Arctech Innovation/LSHTM call centre and Agile Lighthouse (part of NHS Test and Trace). SARS-CoV-2 screening took place in study hospitals, testing centres or remotely through home-testing kits.
Individuals were included in the study if they met all the following criteria: (i) due to have a SARS-CoV-2 swab test or had a test in the previous 72 h, (ii) aged ≥16 years, (iii) had suspected mild or moderate, but not severe, SARS-CoV-2 symptoms and did not require mechanical ventilation or palliative care, or were exposed to SARS-CoV-2 or were an NHS staff member undergoing asymptomatic screening, or a household member of NHS staff, (iv) no previous laboratory confirmed SARS-CoV-2, (v) willing and able to wear a face mask for 3 h, nylon socks and a shirt for 12 h, (vi) provided access to or a copy of their swab test result, and (vii) able and willing to provide written informed consent. Samples from participants who were SARS-CoV-2 RT-PCR positive were placed in the infected sample group and SARS-CoV-2 RT-PCR negative were placed in the uninfected sample group.
Procedures
Samples of breath odours were collected by participants wearing a single-use protective disposable face mask (Fisher Scientific, UK) for 3 h. Skin odours were collected through participants wearing a pair of nylon ankle socks (Retail Premium Grade Try-on Socks, Blue Box Socks, UK) and (t-)shirts, made from natural and synthetic materials, for 12 h. After collection, odour samples were individually wrapped in aluminium foil and packaged in separate labelled polythene bags by the participant. At the point of return, collected samples were stored frozen ≥−20°C.
Participants were followed up on return of samples and 14 days after sample collection, to provide details of ill health. Adverse events and serious adverse events in study participants, staff and dogs were recorded.
Chemical analysis
VOCs from socks collected from 26 asymptomatic or mild symptomatic SARS-CoV-2 infected participants and 27 SARS-CoV-2 uninfected participants were analysed over two days, using a Model 307B VOC analyser (RoboScientific Ltd, UK) fitted with an array of 12 OSC sensors chosen to be sensitive to the VOCs most likely associated with SARS-CoV-2, in this case, ketone and aldehyde compounds25 (Appendix 3 pp 6, available as Supplementary data at JTM online).
Dog training and testing
The methodologies and results for dog training are described in Appendix 3 (pp 6–8) and Appendix 4 (pp 13–14), available as Supplementary data at JTM online, respectively. Here, we summarise the methodology used for double-blind testing.
Sock samples from 200 asymptomatic or mild symptomatic SARS-CoV-2 infected individuals and 200 SARS-CoV-2 uninfected individuals, grouped by sex, age and ethnicity, were used to determine the dogs’ diagnostic accuracy. Samples were generally assigned on a first received basis, but collection site, symptoms on enrolment and condition of samples were also considered. Each sock sample was cut into four pieces, each approximately 80 × 20 mm2, and sealed individually in vented vials (T-mini jar 43 ml, 43 mm diameter, Pattesons Glass Ltd, UK, covered with a clean nylon sock and sealed with a metal cap) to prevent direct contact by the dog and their handler with the samples, and stored at −20°C before use.
Following the training phase, six of seven trained dogs were deemed suitable for the double-blind testing (Appendix 3 pp 8–9, available as Supplementary data at JTM online). Dogs were tested using a three-stand system, each stand holding a glass vial containing a sock sample. Computer software, MDD-Olfactory Performance Recording Application (OPRA), was used to randomly assign odour samples to stands. Therefore, the dogs that worked the three-stand system could have any combination of samples (all infected, all uninfected, or any combination of infected and uninfected) in three positions. A blinded handler, positioned behind a one-way screen (so that the dog could not receive visual prompts from the handler), tasked the dog to search the stands, off lead. Testing required an ‘infected/uninfected’ decision on each sample. Each sample was presented to each dog once, with a maximum of three explorations allowed at the trainer’s discretion.
Statistical analysis
Sample size (Appendix 3 pp 10, available as Supplementary data at JTM online) was based on the requirement to estimate sensitivity and specificity with sufficiently high precision (85 and 90%, respectively). Statistical analyses were done in R version 4.0.3 (chemical analysis) and Stata version 16 (double-blind testing).
Principal components analysis (PCA) was used to identify potential differences in the odours from infected and uninfected individuals, and to obtain biplots (Appendix 3 pp 10, available as Supplementary data at JTM online). In addition, linear discriminant analysis (LDA) was done to determine sensitivity and specificity based on cross-validation. Each day’s evaluation was assessed separately.
Sensitivity and specificity were calculated separately for each dog, assuming PCR was the gold standard. A Bayesian latent class analysis26 that allows for imperfect sensitivity and specificity of PCR was also carried out (Appendix 3 pp 10 and Appendix 4 pp 15, available as Supplementary data at JTM online). The Bayesian analysis had weakly informative priors for the sensitivity and specificity of the dogs and PCR. Improvement in the dogs’ sensitivity and specificity over time and the association with RT-PCR cycle thresholds (Ct) values and sensitivity were assessed using logistic regression with a linear effect for day of study or Ct value, respectively.
Mathematical modelling
We used a modelling approach, adapted from Quilty and co-workers,7 to explore the effectiveness of a ‘Rapid Screen and Test’ strategy using dogs plus confirmatory PCR. We compared this to: (i) a baseline scenario of self-isolation of symptomatic individuals only, (ii) screening with LFTs followed by confirmatory PCR, and (iii) mass screening with PCR (Appendix 3 pp 10, available as Supplementary data at JTM online). We simulated RT-PCR Ct trajectories of infected individuals as a proxy of viral load. Effectiveness was quantified as the proportion of cases identified and the ratio of transmission averted compared to baseline (i.e. isolation of symptomatic individuals only). Transmission averted was calculated as the time that an individual would have spent with a Ct less than 30,27 if they had not been screened and isolated.
Results
Characteristics of study samples
A total of 3921 participants were recruited. A flow chart of participants through the study, summary of reported adverse events and characteristics of samples used in chemical analysis are shown in Appendix 4 pp 11–12, available as Supplementary data at JTM online. Table 1 shows the characteristics of the samples used in double-blind testing.
Table 1.
Characteristics of odour samples used for dog testing
| Infected group (RT-PCR +ve, n = 200) | Uninfected group (RT-PCR −ve, n = 200) | |
|---|---|---|
| Source of sample | ||
| Arctech Innovation/LSHTM call centre and Agile Lighthouse | 175 (87.5%) | 9 (4.5%) |
| NHS hospitals | 25 (12.5%) | 191 (95.5%) |
| Gender | ||
| Women | 147 (73.5%) | 155 (77.5%) |
| Men | 53 (26.5%) | 45 (22.5%) |
| Age, years | ||
| 16–50 | 129 (64.5%) | 117 (58.5%) |
| >50 | 71 (35.5%) | 83 (41.5%) |
| Ethnicity | ||
| White | 190 (95.0%) | 172 (86.0%) |
| Asian | 6 (3.0%) | 4 (2.0%) |
| Black | 1 (0.5%) | 1 (0.5%) |
| Other | 3 (1.5%) | 3 (1.5%) |
| Unknown | 0 (0.0%) | 20 (10.0%) |
| Symptoms at enrolment | ||
| Classic SARS-CoV-2 | 148 (74.0%) | 41 (20.5%) |
| Non-classic SARS-CoV-2 | 52 (26.0%) | 159 (79.5%) |
| Hospital patients | 0 (0.0%) | 0 (0.0%) |
| Symptoms at sample receipt at site | ||
| Classic SARS-CoV-2 | 80 (40.0%) | 11 (5.5%) |
| Non-classic SARS-CoV-2 | 76 (38.0%) | 168 (84.0%) |
| Unknown | 44 (22.0%) | 21 (10.5%) |
| Symptoms after 14 days | ||
| Classic SARS-CoV-2 | 65 (32.5%) | 3 (1.5%) |
| Non-classic SARS-CoV-2 | 121 (60.5%) | 191 (95.5%) |
| Unknown | 14 (7.0%) | 6 (3.0%) |
Symptoms at enrolment, at sample receipt at site and 14-day follow-up were categorised as ‘classic SARS-CoV-2’ if fever, cough, or loss or change of smell or taste were reported, and ‘non-classic SARS-CoV-2’ for those who reported no symptoms or where other symptoms were reported, including, shortness of breath, abdominal pain, muscle and joint pain, conjunctivitis or nausea. NHS hospitals: BHAM (1 uninfected), BSDN (1 infected, 12 uninfected), BUCK (47 uninfected), CAWH (9 uninfected), DBTH (14 infected, 4 uninfected), GETH (1 uninfected), JUHL (3 uninfected), KETG (1 infected, 15 uninfected), KMSF (6 uninfected), MACH (2 infected, 1 uninfected), MCRI (1 infected, 8 uninfected), MGPH (18 uninfected), MYSH (29 uninfected), PGHL (3 infected, 1 uninfected), UCLH (2 infected, 2 uninfected), UHCW (1 infected, 4 uninfected), UHMB (16 uninfected), WHAD (14 uninfected). All swabs were processed through routine NHS channels, apart from 1 positive, which were carried out through non-NHS testing route (private hospital)
Chemical analysis
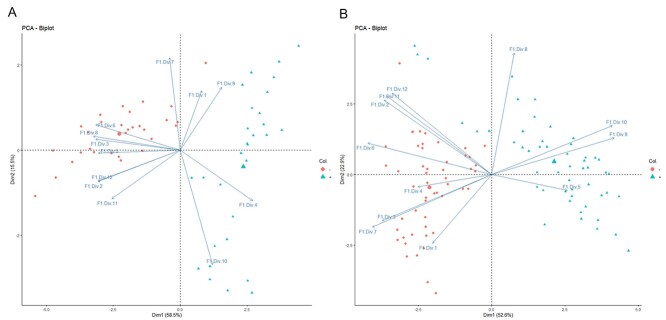
The OSC sensor array was able to distinguish between infected and uninfected samples, demonstrating that SARS-CoV-2 has a distinct odour. On both days of testing, with the first two dimensions, PCA showed clear separation of samples from infected and uninfected participants (Figure 1). OSC sensors achieved 98% (95% CI 95–100) specificity and 99% (95% CI 97–100) sensitivity on the first day of analysis, and 100% sensitivity and specificity on the second.
Figure 1.
Principal component analysis of odour samples by organic semi-conducting (OSC) sensors on two different days; (A) Day 1 and (B) Day 2. Where red circles SARS-CoV-2 infected samples and green triangles are SARS-CoV-2 uninfected odour samples
Double-blind study to assess sensitivity and specificity of dogs
Dogs were able to identify samples from infected (asymptomatic or mild symptoms) individuals with high accuracy. The highest performing dog achieved 94% (95% CI 89–98) sensitivity and 92% (95% CI 88–96) specificity under double-blind conditions (Table 2). Overall, the six dogs achieved a sensitivity range of 82–94% and a specificity range of 76–92%. The positive PPV ranged from 0.4% (95% CI 0.3–0.5) to 55.9% (95% CI 44.3–69.4) and NPV ranged from 98% (95% CI 97–99) to 100% dependent on prevalence rate (Appendix 4 pp 16, available as Supplementary data at JTM online). Specificity (P = 0.003), but not sensitivity (P = 0.650), increased as double-blind testing progressed. There was no evidence for an association between sensitivity and virus quantity in samples, measured by Ct value as a proxy of viral load (P = 0.570, Figure 2A). The range of observed Ct values was 12.0–35.4. The dogs correctly identified B.1.1.7 variant (‘Kent variant’) samples 83% of the time (50/60 presentations). There was no evidence sensitivity to the B.1.1.7 variant was lower (OR = 0.90 [0.45, 1.83], P = 0.777) despite the dogs having only being presented with four samples collected from participants with this variant in training. How sensitivity and specificity was affected by sample characteristics is shown in Appendix 4 pp 16, available as Supplementary data at JTM online.
Table 2.
Sensitivity and specificity for each trained dog (double-blind testing)
| Study group | Analysis assuming PCR as gold standard | Bayesian analysis allowing for imperfect PCR measurements | ||||
|---|---|---|---|---|---|---|
| RT-PCR +ve | RT-PCR −ve | Sensitivity % (95% CI) | Specificity % (95% CI) | Sensitivity % (95% CI) | Specificity % (95% CI) | |
| Asher | 115/129 | 110/132 | 89.1 (82.9–93.6) | 83.3 (76.3–88.9) | 90.9 (85.3–95.4) | 84.8 (77.9–91.1) |
| Kyp | 172/200 | 151/200 | 86.0 (80.7–90.3) | 75.5 (69.2–81.1) | 88.5 (83.6–92.8) | 76.4 (70.3–82.1) |
| Lexi | 172/200 | 165/200 | 86.0 (80.7–90.3) | 82.5 (76.8–87.3) | 90.8 (86.0–94.9) | 85.3 (79.9–90.2) |
| Marlow | 157/200 | 177/200 | 78.5 (72.4–83.8) | 88.5 (83.5–92.4) | 82.1 (76.3–87.3) | 90.1 (85.4–93.9) |
| Millie | 163/200 | 161/200 | 81.5 (75.7–86.4) | 80.5 (74.6–85.5) | 85.5 (80.1–90.5) | 82.6 (76.9–87.6) |
| Tala | 178/200 | 178/200 | 89.0 (84.1–92.8) | 89.0 (84.1–92.8) | 94.3 (89.4–98.0) | 92.0 (87.6–95.8) |
Where data are n/N, CI = confidence intervals
Figure 2.
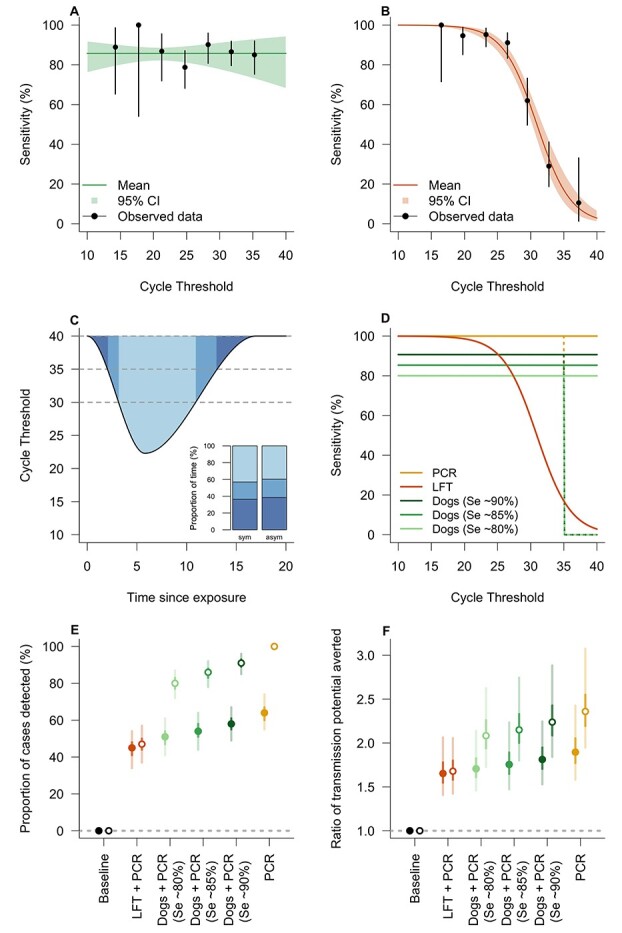
Modelling the effectiveness of a Rapid Screen and Test strategy. The Ct-dependent sensitivity was estimated by fitting a logistic regression model to the results of the double-blind testing (this study) for dogs and to the data presented for the lateral flow test (LFT) in Peto.27 Results show that sensitivity is independent of Ct for dogs (panel A; P = 0.570) whereas sensitivity decreases with increasing Ct values for LFT (panel B; P < 0.0001). The cycle threshold (Ct) is considered a proxy for viral load and is repeatedly simulated from a distribution defined by a starting Ct, a peak Ct and a total duration of infection with a random time since initial exposure. Panel C shows the relationship between Ct and time since exposure for a typical symptomatic individual (asymptomatic individuals having 40% shorter duration of infection). Inset panel shows that both symptomatic and asymptomatic individuals have Ct values between 35 and 40 for approximately one third of the duration of infection. The modelled relationship between sensitivity and Ct for PCR, LFT and dogs is shown in panel D. The sensitivity-Ct relationship for dogs (light green line, 80%; green line, 85%; dark green line, 90%) and LFT (orange line) was informed from data as shown in panels (A) and (B). The sensitivity for PCR was assumed to be 100% up to a Ct of 35, either remaining at this level to a Ct of 40 (yellow solid line) or declining to 0% between 35 and 40 (yellow dotted line). This uncertainty of sensitivity between Ct values of 35 and 40 was also considered for the dogs, with different sensitivity estimated from the data of the double-blind testing (green dotted lines) and representing variability in dog performance. The percentage of cases detected by different strategies is shown in panel E, where baseline corresponds to isolation of symptomatic individuals only and PCR corresponds to the (hypothetical) screening of all individuals with PCR. LFT + PCR and Dogs + PCR indicate, respectively, rapid mass screening with LFTs or dogs followed by confirmatory PCR of positively identified cases. The ratio of the transmission averted by these scenarios compared to baseline is shown in panel F. In panels E and F, filled and open points correspond to a Ct detection limit of 35 and 40 respectively
Mathematical modelling
Modelling indicated that a strategy using dogs, plus RT-PCR for those people indicated as positive by dogs, detected 89% (95% credible interval: 82–95%; Figure 2E) of cases resulting in 2.21 (95% credible interval: 1.83–2.85; Figure 2F) times as much transmission averted compared to isolation of symptomatic individuals only. In comparison, mass testing with RT-PCR alone detected 100% of cases and the amount of transmission averted was 2.37 (95% credible interval: 1.96–3.11), demonstrating the performance of dogs was similar to RT-PCR. Screening using dogs was superior to using LFTs for all assumptions on dog sensitivity (varied between 80 and 90%) but is dependent on the assumed sensitivity of RT-PCR for low viral loads (Figure 2E and F). The sensitivity of RT-PCR at low viral loads, as occurs during the early or late stage of infection or if true asymptomatic, is not well established in the literature, and therefore, we modelled all testing scenarios (LFT + RT-PCR, dogs + RT-PCR and RT-PCR alone) with 100% sensitivity of RT-PCR up to either 35 or 40 Ct for comparison. It is noteworthy that if RT-PCR has, in practice, no sensitivity in the Ct 35–40 range, even mass testing using RT-PCR would detect only approximately 64% of infections (Figure 2E). This is because infected individuals have a Ct value between 35 and 40 for approximately one third of the duration of infection (Figure 2C, inset).
Discussion
Principal findings
This study demonstrated that there is a distinct body odour associated with asymptomatic and mild symptomatic SARS-CoV-2 infections, and that OSC sensors and trained dogs are able to identify this odour with a high degree of accuracy. After just six weeks training, six dogs discriminated between odour samples from 200 infected participants and 200 uninfected participants with a sensitivity range of 82–94% and a specificity range of 76–92% compared with the reference test, RT-PCR. In our analysis, we adjusted for the RT-PCR being imperfect and recognised that there was a degree of concordance between dogs. Modelling showed that trained bio-detection dogs could be used at ports-of-entry or other sites with large numbers of people and should be considered as a new rapid screening tool.
Meaning of the study
Work with the sensors shows that the VOCs associated with SARS-CoV-2 can generate an odour ‘fingerprint’ as the sensors were tuned to ketone and aldehyde compounds; therefore, the development of an OSC sensor device, which could be used to screen air from rooms (e.g. classrooms) or aircraft cabins could be developed. This would allow the detection of one infected individual within a room or aircraft, allowing rapid and more targeted testing to be done, saving money and time, and reducing onward transmission. The confirmation by VOC analysis that there is a distinct odour between the two groups, and the addition of the tentative identification of the volatile chemicals involved, may also enable the production of training aids for dogs (pseudo-odours), reducing the time spent required to obtain samples for training.
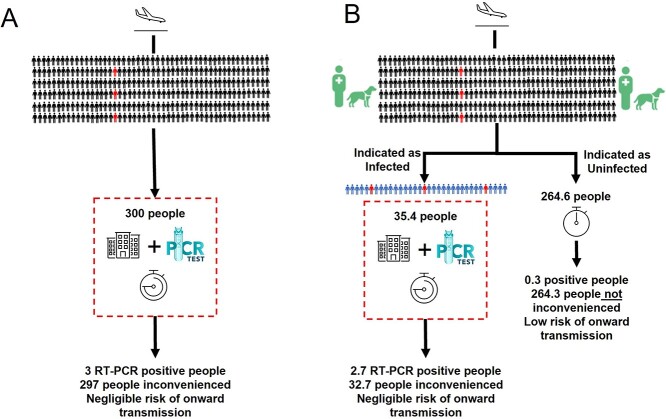
The bio-detection dogs, in this experimental setting, had a higher accuracy than the LFT, which has a wider range and lower overall sensitivity of between 58 and 77%.28 RT-PCR is the gold standard test due to a high sensitivity (97–99%) and specificity (95–99%),29 but dogs have a major advantage over both these tests as they are incredibly rapid. Our preparatory work indicates that two dogs could screen 300 people in 30 min, for example, the time it takes to disembark from a plane, and PCR would only need to be used to test those individuals identified as positive by the dogs (Figure 3). This would result in far fewer individuals needing an RT-PCR test, allowing most travellers to continue their onward journey or mass event attendees with little inconvenience. If used at airports, dogs may also serve as a visual deterrent, reducing passengers travelling with false SARS-CoV-2 negative certificates. A relatively narrow range of sensitivity and specificity was apparent between the different dogs tested, but in practice, only the highest performing dogs would be deployed. Specificity also improved during testing, and we postulate that both will improve further in real-world settings, with SARS-CoV-2-positive passengers providing larger and clearer odour profiles than used for this study.
Figure 3.
Exemplar of (A) Current SARS-CoV-2 Strategy for red list countries and unvaccinated travellers (10-day quarantine and PCR tests) and (B) Proposed Rapid Screen and Test Strategy. Schematic outlining the number of true negatives (black) and true positives (red) and false negatives (blue) as a result of screening people, with 1% SARS-CoV-2 prevalence, followed by confirmatory PCR testing. Assuming 100% sensitivity and specificity of RT-PCR, and 90% sensitivity and 89% specificity of dogs (values used in the mathematical modelling). ‘Inconvenienced’ refers to virus-negative passengers required to be in quarantine (red dotted line)
Mathematical modelling suggested that a ‘Rapid Screen and Test’ strategy, using dogs plus confirmatory real-time RT-PCR of positively identified individuals, could be highly effective in detecting cases and averting transmission. This screening strategy could be used in a variety of targeted settings and scenarios where the greatest impact could be achieved. Our results indicate that dogs outperform LFT (as an alternative rapid screening tool) across sensitivities between 80 and 90%. An important reason for this is the seeming independence of dog sensitivity and viral load (using Ct as a proxy), which contrasts with the rapid decline in sensitivity for the LFT with increasing Ct. Even if the sensitivity of dogs fell to zero for Ct values greater than 35, they would still perform better than LFTs, due to the extremely low sensitivity of LFTs for Ct values in this range. Interestingly, we found our estimates of effectiveness to be very sensitive to the performance of RT-PCR in detecting low viral loads (Ct values > 35). For example, in the best-case scenario where RT-PCR sensitivity remains high for Ct > 35, the Rapid Screen and Test strategy detects 80–90% of cases. This drops precipitously to 50–60% if the diagnostics cannot (in practice) detect Ct values between 35 and 40, indicative of low viral loads. Indeed, that RT-PCR alone can detect only approximately 60% of infections if it is insensitive in the Ct 35–40 range, may have implications for the interpretation of prevalence estimates made by random testing of populations. The proportion of time an infected individual spends with low Ct in the 35–40 range is similar for symptomatic and asymptomatic individuals,28 indicating that adjusting the ratio of asymptomatic and symptomatic individuals will have little impact on the projected performance of Rapid Screen and Test. In future, the modelling approach could be used to evaluate the effectiveness of screening in other contexts, such as public venues, mass events and domestic travel hubs where case detection could avert potential superspreading events.
Strengths and limitations
We have shown that OSC sensors can detect changes in body odour associated with SARS-CoV-2. It is also the first study to assess whether trained dogs can distinguish between the odour of people infected with SARS-CoV-2 and those who are uninfected, in a randomized double-blind trial, where trainer and monitor were unaware of the study group for each sample, and with a sufficiently high number of trained dogs tested and individuals donating samples. We are confident that the dogs identified a specific odour signature associated with infection with SARS-CoV-2, as they were tested double blind on samples that had not been used during training. Similar values of sensitivity and specificity have been recorded in a number of pilot studies20–25 although uncertainty in the estimates from these studies is high because of weak study designs. The common limitations of these studies include an insufficient sample size, samples collected from only hospitalised patients, and excluding uninfected people with cold-like symptoms. Also, limitations of dog training included the use of a positive specimen in each test line, so the dog identifies the ‘odd’ sample (known as a forced choice paradigm), and not testing the dogs on novel samples in a double-blind fashion. Unlike in most previous studies, we included asymptomatic and mild symptomatic cases of SARS-CoV-2 and demonstrate that dogs can identify these individuals, including some with extremely low virus titres, as suggested by high Ct values using real-time RT-PCR.
Our study has a number of limitations. Firstly, although sensors and dogs could be used to screen samples, the real value would be screening people. Our recent on-going work shows that dogs trained with t-shirts collected in this study readily transition from laboratory-sample testing to identifying people infected with SARS-CoV-2 (Appendix 5 pp 17–20, available as Supplementary data at JTM online). This work is encouraging and suggests that trained dogs will readily identify people infected with SARS-CoV-2 from lines of uninfected people. Future work is also required to understand the robustness of the sensors as only a limited sample size was used in this present study, as well as the VOC concentration required for sensor detection. Although the preliminary work with the dogs and t-shirts is also encouraging for the use of sensors in a real-life situation, as both were developed using the same odour. Secondly, our results suggest dogs are able to detect the B.1.1.7 variant, although the sample size was not sufficient for a reliable estimate of sensitivity of this variant. In the event that a new variant of SARS-CoV-2 resulted in a different odour profile, trained dogs could be rapidly re-trained to detect the new odour within two days providing odour samples for the new variant are available. Thirdly, there is a possibility that other respiratory viral infections produce similar odour signatures to SARS-CoV-2. This is, however, unlikely given that 26% of PCR-confirmed uninfected participants in our study displayed classic SARS-CoV-2, cold or flu-like symptoms, and were correctly identified as uninfected. Additionally, other studies suggest that different viral infections result in distinct odour profiles.8–11
Conclusion
Our study demonstrated that trained dogs and OSC sensors can detect people with asymptomatic and mild SARS-CoV-2 infections by their odour with a high degree of accuracy under laboratory conditions and should be considered as an additional tool for use in SARS-CoV-2 testing strategies, used in conjunction with a confirmatory PCR to confirm those individuals indicated as positive by dogs. Dogs should be considered for deployment alongside LFTs for certain scenarios as a more rapid screening method to detect individuals who are most likely to be infectious, and further modelling work on this strategy is warranted. Overall, using COVID detection dogs as a screening strategy is the quickest, non-invasive method with high accuracy and would profoundly improve our ability to screen large numbers of people for COVID-19.
Dissemination Declaration
All participants in this study will be emailed a newsletter containing the results of this study.
Data Sharing
All data will be archived at Arctech Innovation. A dataset containing de-identified participant data that support the findings of this article is available from the corresponding author (JL) upon reasonable request and accompanied by IRB approval. Data is available immediately, for 10 years, after, which archives may be destroyed. The study protocol is in Appendix 6, available as Supplementary data at JTM online.
Supplementary Material
Acknowledgements
We are grateful to the participants who donated the samples, staff in NHS hospitals and testing centres. We thank members of the Trial Steering Committee for their helpful suggestions, time and commitment. Electronic data solutions were provided by LSHTM Global Health Analytics (odk.lshtm.ac.uk).
Contributor Information
Claire Guest, Medical Detection Dogs, Milton Keynes, UK.
Sarah Y Dewhirst, Arctech Innovation, The Cube, Londoneast-uk Business and Technical Park, Dagenham, UK.
Steve W Lindsay, Department of Biosciences, Durham University, Durham, UK.
David J Allen, Department of Infection Biology, Faculty of Infectious and Tropical Diseases, London School of Hygiene and Tropical Medicine, London, UK.
Sophie Aziz, Medical Detection Dogs, Milton Keynes, UK.
Oliver Baerenbold, Department of Medical Statistics, London School of Hygiene and Tropical Medicine, London, UK.
John Bradley, MRC International Statistics and Epidemiology Group, London School of Hygiene and Tropical Medicine, London, UK.
Unnati Chabildas, Arctech Innovation, The Cube, Londoneast-uk Business and Technical Park, Dagenham, UK.
Vanessa Chen-Hussey, Arctech Innovation, The Cube, Londoneast-uk Business and Technical Park, Dagenham, UK.
Samuel Clifford, Department of Infectious Disease Epidemiology, Faculty of Epidemiology and Population Health, London School of Hygiene and Tropical Medicine, London, UK.
Luke Cottis, Hampden Veterinary Hospital, Anchor Ln, Aylesbury, UK.
Jessica Dennehy, Arctech Innovation, The Cube, Londoneast-uk Business and Technical Park, Dagenham, UK.
Erin Foley, Arctech Innovation, The Cube, Londoneast-uk Business and Technical Park, Dagenham, UK.
Salvador A Gezan, Arctech Innovation, The Cube, Londoneast-uk Business and Technical Park, Dagenham, UK.
Tim Gibson, RoboScientific Ltd, Ely, UK.
Courtenay K Greaves, Arctech Innovation, The Cube, Londoneast-uk Business and Technical Park, Dagenham, UK.
Immo Kleinschmidt, MRC International Statistics and Epidemiology Group, London School of Hygiene and Tropical Medicine, London, UK.
Sébastien Lambert, Royal Veterinary College, University of London, Hatfield, UK.
Anna Last, Clinical Research Department, Faculty of Infectious and Tropical Diseases, London School of Hygiene and Tropical Medicine, London, UK.
Steve Morant, Medical Detection Dogs, Milton Keynes, UK.
Josephine E A Parker, Arctech Innovation, The Cube, Londoneast-uk Business and Technical Park, Dagenham, UK.
John Pickett, Cardiff University Main Building, Cardiff, UK.
Billy J Quilty, Department of Infectious Disease Epidemiology, Faculty of Epidemiology and Population Health, London School of Hygiene and Tropical Medicine, London, UK.
Ann Rooney, Lomond Veterinary Clinic, Helensburgh, UK.
Manil Shah, Arctech Innovation, The Cube, Londoneast-uk Business and Technical Park, Dagenham, UK.
Mark Somerville, Medical Detection Dogs, Milton Keynes, UK.
Chelci Squires, Arctech Innovation, The Cube, Londoneast-uk Business and Technical Park, Dagenham, UK.
Martin Walker, Royal Veterinary College, University of London, Hatfield, UK.
James G Logan, Arctech Innovation, The Cube, Londoneast-uk Business and Technical Park, Dagenham, UK; Department of Disease Control, Faculty of Infectious and Tropical Diseases, London School of Hygiene and Tropical Medicine, London, UK.
COVID Dogs Research Team:
Robert Jones, Ana Assis, Ewan Borthwick, Laura Caton, Rachel Edwards, Janette Heal, David Hill, Nazifa Jahan, Cecelia Johnson, Angela Kaye, Emily Kirkpatrick, Sarah Kisha, Zaena Ledeatte Williams, Robert Moar, Tolulope Owonibi, Benjamin Purcell, Christopher Rixson, Freya Spencer, Anastasios Stefanidis, Sophie Stewart, Scott Tytheridge, Sian Wakley, Shanice Wildman, Catherine Aziz, Helen Care, Emily Curtis, Claire Dowse, Alan Makepeace, Sally-Anne Oultram, Jayde Smith, Fiona Shenton, Harry Hutchins, Robert Mart, Jo-anne Cartwright, Miranda Forsey, Kerry Goodsell, Lauren Kittridge, Anne Nicholson, Angelo Ramos, Joanne Ritches, Niranjan Setty, Mark Vertue, Malin Bergstrom, Zain Chaudhary, Angus De Wilton, Kate Gaskell, Catherine Houlihan, Imogen Jones, Marios Margaritis, Patricia Miralhes, Leah Owens, Tommy Rampling, Hannah Rickman, Marta Boffito, Candida Fernandez, Bryony Cotterell, Anne-Marie Guerdette, George Tsaknis, Margaret Turns, Joanne Walsh, Lisa Frankland, Raha West, Maureen Holland, Natalie Keenan, Helen Wassall, Megan Young, Jade Rangeley, Gwendolyn Saalmink, Sanjay Adlakha, Philip Buckley, Lynne Allsop, Susan Smith, Donna Sowter, Alison Campbell, Julie Jones, Steve Laird, Sarah O’Toole, Courteney Ryan, Jessica Evans, James Rand, Natasha Schumacher, Tracey Hazelton, Andrew Dodgson, Susannah Glasgow, Denise Kadiu, Orianne Lopuszansky, Anu Oommen, Joshi Prabhu, Molly Pursell, Jane Turner, Hollie Walton, Robert Andrews, Irena Cruickshank, Catherine Thompson, Tania Wainwright, Alun Roebuck, Tara Lawrence, Kimberley Netherton, Claire Hewitt, Sarah Shephardson, Winston Andrew Crasto, Judith Lake, Rosemary Musanhu, Rebecca Walker, Karen Burns, Andrew Higham, Julie Le Bas, Nicola Mackenzie, Hilary Thatcher, Shannen Beadle, Sarah Buckley, Gail Castle, Aimee Fletcher, Sara Holbrook, Patricia Kane, Kate Lindley, Tracey Lowry, Stephanie Lupton, Sharon Oddy, Lynda Slater, Martin Sylvester, Kenneth Agwuh, Veronica Maxwell, Stephen Ryder, Kirsty Topham, Obi Egbuniwe, Rebecca Matthews, Alejandro Arenas-Pinto, Paulina Prymas, Abigail Severn, Amber Shaw, Safia Begum, Daniel Lenton, James Scriven, Lucy Leeman, Karen Rudge, Emma Storr, Ana Alvarez, Kate Forster, Daniel Hind, Natalie Cook, Rosanna Peeling, Peter Carey, Anne Wilson, and Jane Davis
Author Contributors
J.L., C.G., S.W.L., A.L., J.B., I.K., D.J.A., S.Y.D. conceived and designed the study. I.K., J.B. planned the statistical analysis. J.L. led the study. S.Y.D., C.S., U.C., M.S. oversaw collection of samples from NHS hospitals. S.M., S.A. generated the random allocation sequence that assigned the samples for testing. S.Y.D., J.E.A.P., E.F. supervised collection, collation, preparation and storage of samples. S.Y.D., U.C., M.S. implemented quality assurance for the trial. A.L., C.K.G. took responsibility for patients in the trial and A.R., L.C. for the health of the dogs. C.G., M.S., S.A. trained and tested the dogs. T.G. screened SARS-CoV-2 VOCs and produced the OSC sensor array. T.G., J.P. carried out the chemical analysis. V.C.H. led the data management team. J.B., O.B., S.A.G., T.G. did the statistical analysis. B.J.Q., S.C., M.W., S.L., J.D. performed the mathematical modelling. S.W.L., S.Y.D., J.L., E.F., J.D. produced the first draft of the manuscript. All authors read and approved the final manuscript. J.L. is a guarantor. The corresponding author attests that all listed authors meet authorship criteria and that no others meeting the criteria have been omitted.
Funding
The study was supported by a grant from Department of Health and Social Care, UK Government (2020/023), Durham University COVID-19 response fund (RF020929), NIHR Clinical Research network support (IRAS ID 284222) and charitable donations.
Conflict of interest
Medical Detection Dogs is a registered charity in England and Wales No. 1124533 and in Scotland No. SC044434. Tim Gibson is the Chief Scientific Officer of RoboScientific and holds shares. All remaining authors have declared no conflicts of interest.
References
- 1. Hafeez S, Din M, Zia F et al. Emerging concerns regarding COVID-19; second wave and new variant. J Med Virol 2021; 93:4108–4110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Mahase E. Covid-19: what new variants are emerging and how are they being investigated? BMJ 2021; 372:n158. [DOI] [PubMed] [Google Scholar]
- 3. Gostic K, Gomez ACR, Mummah RO et al. Estimated effectiveness of symptom and risk screening to prevent the spread of COVID-19. Elife 2020; 9:e55570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Mina MJ, Peto TE, García-Fiñana M et al. Clarifying the evidence on SARS-CoV-2 antigen rapid tests in public health responses to COVID-19. Lancet 2021; 397:1425–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Lee LYW, Rozmanowski S, Pang M et al. SARS-CoV-2 infectivity by viral load, S gene variants and demographic factors and the utility of lateral flow devices to prevent transmission. Clin Infect Dis 2021; 74:407–415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Pickering S, Batra R, Snell LB et al. Comparative performance of SARS-CoV-2 lateral flow antigen tests and association with detection of infectious virus in clinical specimens: a single-centre laboratory evaluation study. Lancet Microbe 2021; 2:e461–e471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Quilty BJ, Clifford SC, Hellewell J et al. Quarantine and testing strategies in contact tracing for SARS-CoV-2: a modelling study. Lancet Public Health 2021; 6:E175–E183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. El Qader AA, Lieberman D, Avni YS et al. Volatile organic compounds generated by cultures of bacteria and viruses associated with respiratory infections. Biomed Chromatogr 2015; 29:1783–90. [DOI] [PubMed] [Google Scholar]
- 9. Purcaro G, Rees CA, Wieland-Alter WF et al. Volatile fingerprinting of human respiratory viruses from cell culture. J Breath Res 2018; 12:026015–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Schivo M, Aksenov AA, Linderholm AL et al. Volatile emanations from in vitro airway cells infected with human rhinovirus. J Breath Res 2014; 8:037110–0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Mashir A, Paschke KM, van Duin D et al. Effect of the influenza A (H1N1) live attenuated intranasal vaccine on nitric oxide (FENO) and other volatiles in exhaled breath. J Breath Res 2011; 5:037107. 10.1088/1752-7155/5/3/037107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Jones RT, Guest C, Lindsay SW et al. Could bio-detection dogs be used to limit the spread of COVID-19 by travellers? J Travel Med 2020; 27:taaa131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Ehmann R, Boedeker E, Friedrich U et al. Canine scent detection in the diagnosis of lung cancer: revisiting a puzzling phenomenon. Eur Respir J 20121; 39:669–76. [DOI] [PubMed] [Google Scholar]
- 14. Rooney NJ, Morant S, Guest C. Investigation into the value of trained glycaemia alert dogs to clients with type I diabetes. PLoS One 2013; 8:e69921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Willis CM, Church SM, Guest CM et al. Olfactory detection of human bladder cancer by dogs: proof of principle study. BMJ 2004; 329:712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Guest C, Pinder M, Doggett M. Trained dogs identify people with malaria parasites by their odour. Lancet Infect Dis 2019; 19:578–80. [DOI] [PubMed] [Google Scholar]
- 17. Walker D, Walker J, Cavnar P et al. Naturalistic quantification of canine olfactory sensitivity. Appl Anim Behav Sci 2006; 97:241–54. [Google Scholar]
- 18. Vesga O, Valencia AF, Mira A et al. Dog savior: immediate scent-detection of SARS-COV-2 by trained dogs. bioRxiv 2020.06.17.158105; doi: 10.1101/2020.06.17.158105. [DOI] [Google Scholar]
- 19. Grandjean D, Sarkis R, Lecoq-Julien C et al. Can the detection dog alert on COVID-19 positive persons by sniffing axillary sweat samples? A proof-of-concept study. PLoS ONE 2020; 15:e0243122. https://doi.org/10.1371/journal. pone.0243122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Jendrny P, Schulz C, Twele F et al. Scent dog identification of samples from COVID-19 patients–a pilot study. BMC Infect Dis 2020; 20:1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Jendrny P, Twele F, Meller S et al. Scent dog identification of SARS-COV-2 infections, similar across different body fluids. BMC Infect Dis 2021; 21:707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Eskandari E, Marzaleh MA, Roudgari H et al. Sniffer dogs as a screening/diagnostic tool for COVID-19: a proof of concept study. BMC Infect Dis 2021; 21:243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Granjean D, Al Marzooqi DH, Lecoq-Julien C et al. Use of canine olfactory detection for COVID-19 testing study on UAE trained detection dog sensitivity. J Vet Sci Res 2021; 6:000210. [Google Scholar]
- 24. Essler JL, Kane SA, Nolan P et al. Discrimination of SARS-CoV-2 infected patient samples by detection dogs: a proof of concept study. PLoS One 2021; 16:e0250158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Ruszkiewicz DM, Sanders D, O’Brien R et al. Diagnosis of COVID-19 by analysis of breath with gas chromatography-ion mobility spectrometry—a feasibility study. EClinicalMedicine 2020; 3329-30:100609–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Dendukuri N, Joseph L. Bayesian approaches to modeling the conditional dependence between multiple diagnostic tests. Int Biometric Soc 2001; 57:158–67. [DOI] [PubMed] [Google Scholar]
- 27. Peto T. UK COVID-19 lateral flow oversight team. COVID-19: rapid antigen detection for SARS-CoV-2 by lateral flow assay: a national systematic evaluation for mass-testing. E Clinical Medicine 2021; 36:100924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Oxford University and PHE Porton Down. University of Oxford SARS-CoV-2 test development and validation cell. Preliminary report from the Joint PHE Porton Down & University of Oxford SARS-CoV-2 test development and validation cell: Rapid evaluation of Lateral Flow Viral Antigen detection devices (LFDs) for mass community testing; 2020. www.ox.ac.uk/sites/files/oxford/media_wysiwyg/UK%20evaluation_PHE%20Porton%20Down%20%20University%20of%20Oxford_final.pdf
- 29. Tsang NNY, So HC, Ng KY et al. Diagnostic performance of different sampling approaches for SARS-CoV-2 RT-PCR testing: a systematic review and meta-analysis. Lancet Infect Dis 2021; 21:1233–1245. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.