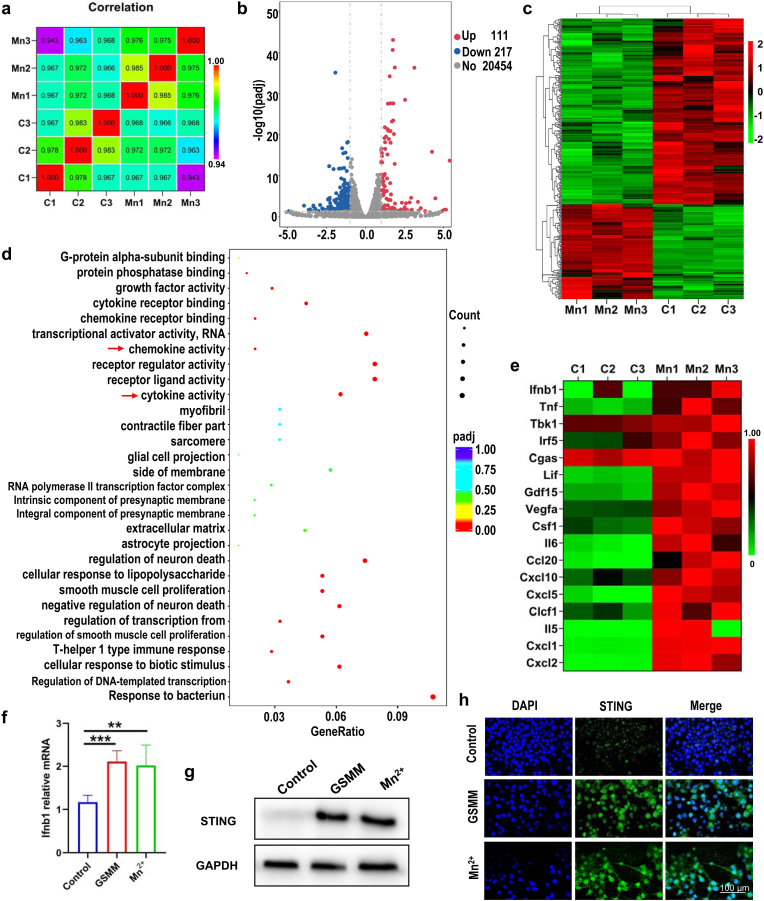
Fig. 3.
The excavation of biological mechanism of GSMM by RNA sequencing. (a) Sample correlation test. (b) The volcano map of GSMM (n = 3: Mn1, Mn2, Mn3) and control (n = 3: C1, C2, C3) group. (c) The heatmap of genes alteration after GSMM treatment (p < 0.05, [fold change≥2]). (d) Gene ontology enriched in GSMM treatment and control group (red arrows represent GSMM relative pathways. (e) The heatmap of selected genes related to cGAS-STING signaling pathway. (f) The relative mRNA expression of ifnb1 was detected by qRT-PCR (One-way ANOVA, **: p < 0.01, ***: p < 0.001). (g) The western blotting of STING in GSMM, MnCl2 and control group. (h) Immunofluorescence images of STING in GSMM, MnCl2 and control group. Scale bar: 100 μm.