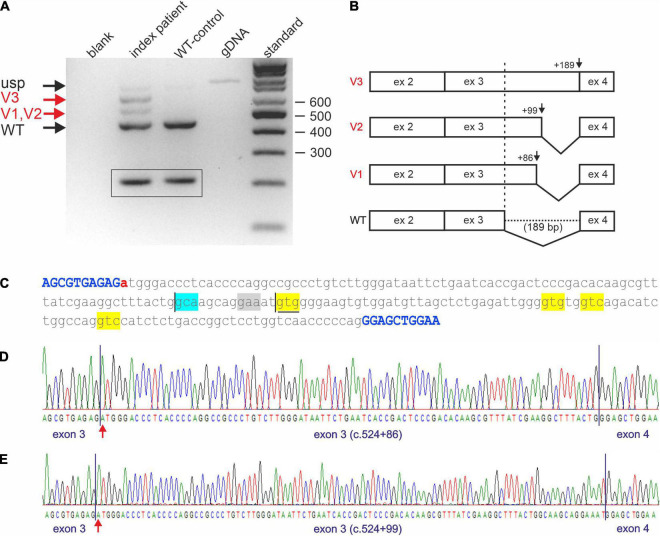
FIGURE 2.
Impaired STUB1 splicing. (A) Expression studies of processed STUB1 transcripts of the index patient (II-4) and a control. (blank) control without template, (index patient) reverse transcription polymerase chain reaction (RT-PCR) on RNA of the index patient, (WT-control) RT-PCR on RNA of the WT-control, (gDNA) genomic DNA, (standard) 100 bp size standard. Black arrows indicate the correctly processed exon 2–4 fragment of STUB1 (WT) and the weak, unspliced (usp) product. Red arrows indicate incorrect splicing in the index patient. Splice variants 1, and 2 (V1, V2) correspond to the cryptic splice sites, while splice variant 3 (V3) contains retained intron 3 of STUB1. TBP-derived 210 bp control fragments for RNA integrity are shown in boxes. (B) Graphical overview of impaired splicing (V3-V1) caused by variant c.524+1G>A compared to the correctly spliced wild type (WT) allele. (C) Sequence of intron 3 of STUB1 (lower case) flanked by exons 3 and 4 (upper case, blue). Pathogenic variant c.524+1G>A is highlighted in red. The main cryptic splice site c.524+86 is boxed in blue. Predicted cryptic splice donor sites by Alternative Splice Site Predictor analysis are boxed in yellow, the predicted splice acceptor is boxed in gray. The ultra-rare used cryptic splice site c.524+99 is underlined. (D) Electropherogram of products using the cryptic splice site c.524+86. (E) Electropherogram of products using the ultra-rare cryptic splice site c.524+99.