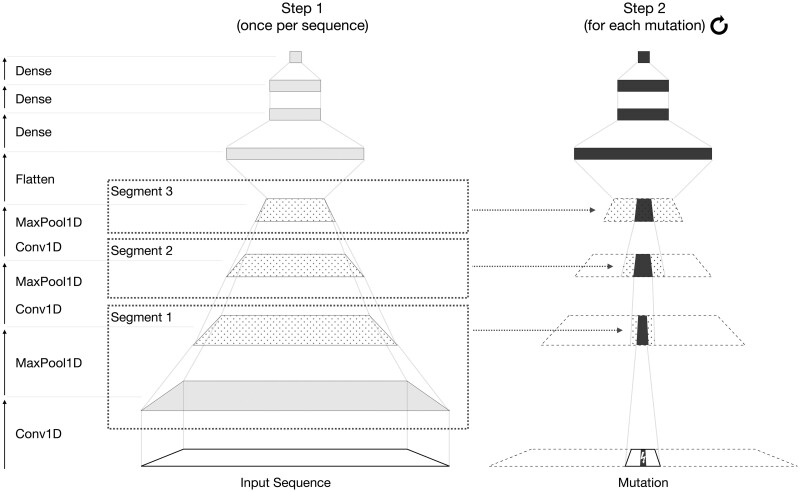
Fig. 3.
Overview of the fastISM algorithm for an input model similar to the Basset-like architecture in Figure 2. fastISM inspects the input model and partitions successive convolution and maxpooling layers into segments. For each input sequence, the input model is run once (Step 1) and returns the intermediate outputs of the final layer of each segment (dotted), which is cached. Note that after the first Conv1D and MaxPool1D layers, the subsequent Conv1D and MaxPool1D layers are shown as a single operation; similar to Segment 1, only the output of the maxpooling layer is cached. For each mutation of each sequence (Step 2), cached values are sliced appropriately, and recomputed values (dark filled-in) are overlaid at each layer. We omit layers, such as activations and batch normalization here for simplicity