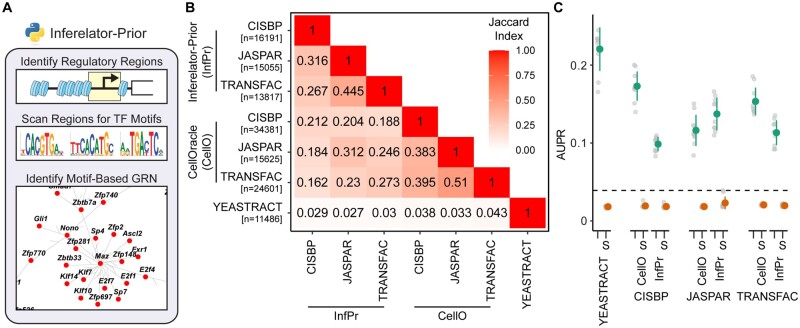
Fig. 3.
Construction and performance of network connectivity priors using TF motif scanning. (A) Schematic of inferelator-prior workflow, scanning identified regulatory regions (e.g. by ATAC) for TF motifs to construct adjacency matrices. (B) Jaccard similarity index between S.cerevisiae prior adjacency matrices generated by the inferelator-prior package, by the CellOracle package, and obtained from the YEASTRACT database. Prior matrices were generated using TF motifs from the CIS-BP, JASPAR and TRANSFAC databases with each pipeline (n is the number of edges in each prior adjacency matrix). (C) The performance of Inferelator network inference using each motif-derived prior. Performance is evaluated by AUPR, scoring against genes held out of the prior adjacency matrix, based on inference using 2577 genome-wide microarray experiments. Experiments labeled with (S) are shuffled controls, where the labels on the prior adjacency matrix have been randomly shuffled. The black dashed line is the performance of the GRNBOOST2 algorithm, which does not incorporate prior knowledge, scored against the entire gold standard network