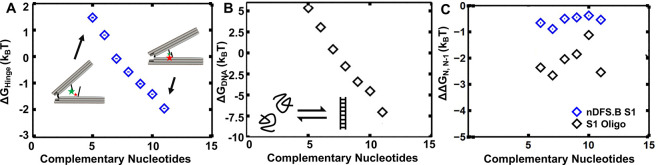
Figure 2.
(A) Plot of ΔGhingevs number of complementary bases for nDFS.B S1, computed using the Boltzmann weight of the probability of the closed state as measured by the FRET ensemble data. (B) Plot of ΔGDNA, the difference in free energy between the S1 DNA melted and annealed states vs length of complementary sequence computed using salt-adjusted free energies33,34 with a reference concentration of 1 μM. (C) ΔΔGN,N–1 computed separately from (A) ΔGhinge or (B) ΔGDNA, where ΔΔGN,N–1 is the difference between ΔGN and ΔGN–1, which is the additional stability imparted by the addition of one base pair. The lower ΔΔGN,N–1 for the DNA alone relative to the nDFS.B implies that the device acts on the oligonucleotides to alter the free energy of binding.