Figure 2.
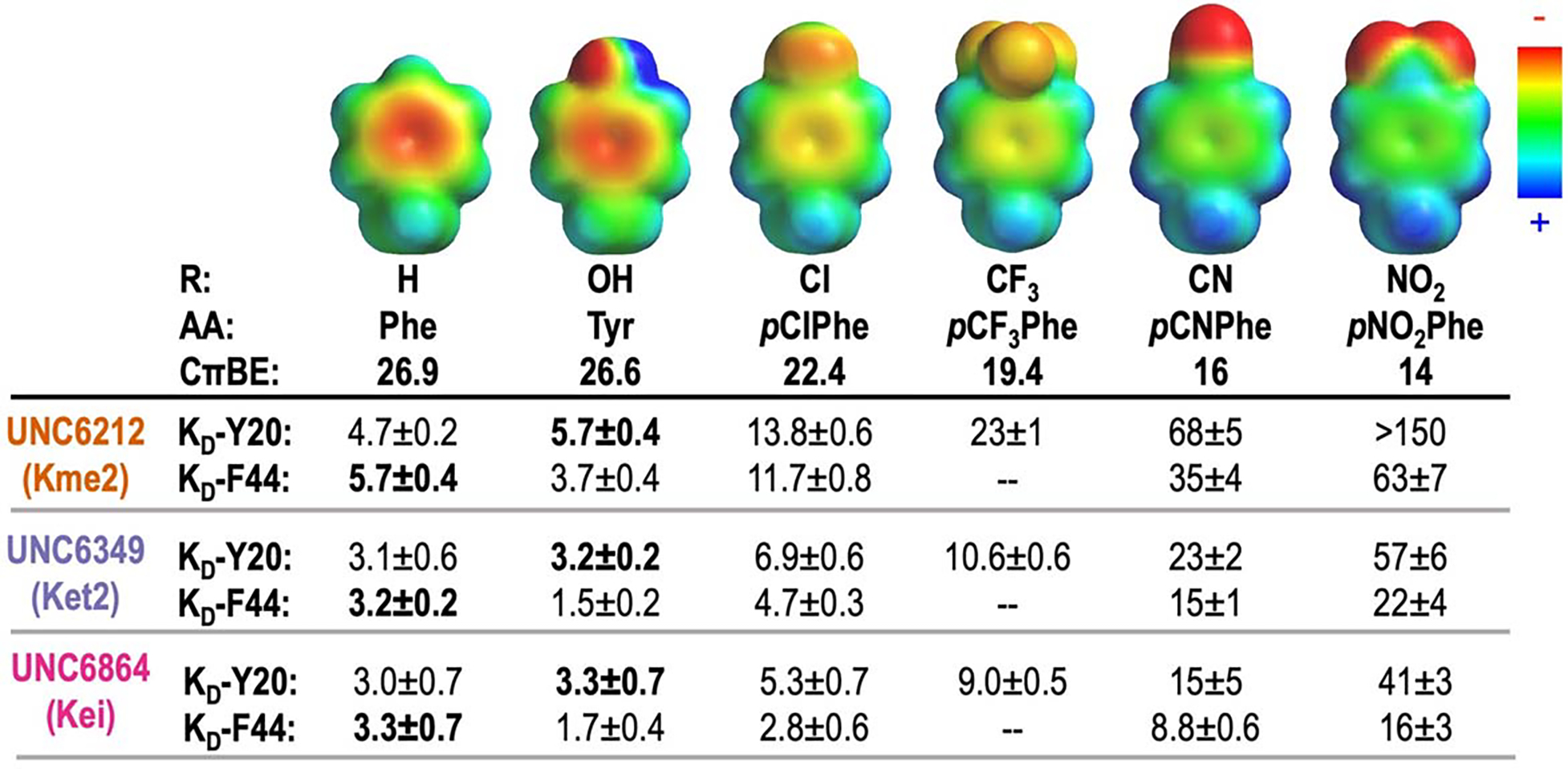
The effect of R-groups on electrostatic surface potential (ESP) maps and calculated cation-π binding energy (CπBE, kcal/mol) of amino acids tested within the aromatic cage. ESP maps were calculated in Spartan at the ωB97X-D/6–31G(d) level of theory. CπBE are calculated for substituted benzenes and Na+.20 Measured KD values (μM) for CBX5 variants at positions Y20 and F44 with peptide-based ligands, UNC6212 (Kme2), UNC6349 (Ket2), and UNC6864 (Kei). Affinities corresponding to wild-type protein are shown in bold. Binding affinity of the F44pCF3Phe variant was not determined. Errors given for KD are the standard deviation for 3 experimental replicates or the highest individual error from an individual experiment among the replicates, whichever is greater.
