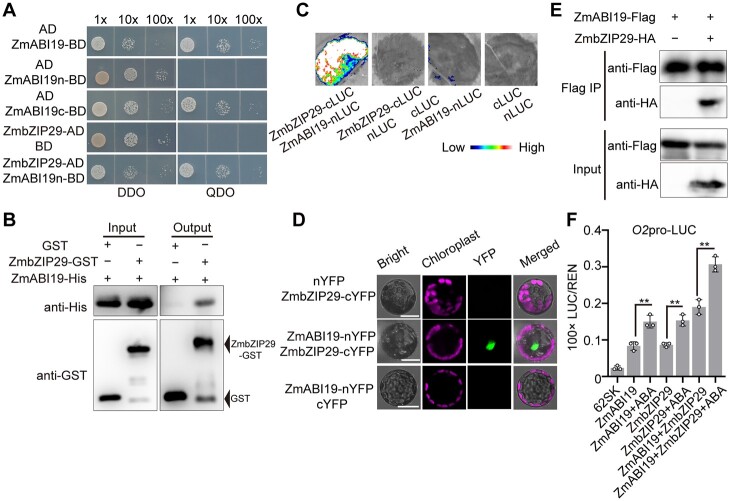
Figure 4.
Synergistic transactivation of the O2 promoter by ZmABI19, ZmbZIP29, and ABA. A, Y2H assay showing the interaction between ZmABI19 and ZmbZIP29. The full-length region (1–293 amino acid), N-terminus (1–72 amino acid), and C-terminus (73–293 amino acid) of ZmABI19 were cloned into the pGBKT7 plasmid, yielding ZmABI19-BD, ZmABI19n-BD, and ZmABI19c-BD. The CDS of ZmbZIP29 was cloned into pGADT7, yielding ZmbZIP29-AD. Serial dilutions (1:1, 1:10, 1:100) of transformed yeast cells were grown on the SD/−Leu/−Trp (DDO) and SD/−Leu/−Trp/−His/−Ade (QDO) medium. B, In vitro pull-down assay showing the interaction between ZmABI19 and ZmbZIP29. C, LIC assay showing the interaction between ZmABI19 and ZmbZIP29. The LUC intensity was measured through co-expression of different combinations. Low, weak LUC intensity; High, strong LUC intensity. D, BiFC assay showing the interaction between ZmABI19 and ZmbZIP29. Bars = 50 μm. E, Co-IP showing the interaction between ZmABI19 and ZmbZIP29. Total proteins were extracted from N. benthamiana leaves, where the ZmbZIP29-HA and ZmABI19-Flag fusion proteins were co-expressed. The ZmABI19-Flag proteins were immunoprecipitated with Flag agarose beads and detected with anti-HA and anti-Flag antibodies, respectively. F, Transactivation of the O2 promoter by ZmABI19, ZmbZIP29, and ABA. 10-μM ABA was used in this assay. The relative ratio of LUC/REN was determined in Arabidopsis protoplasts via co-transforming the reporter plasmids with the effector construct. Error bars represent SD of three biological replicates. Statistical significance was determined with Student’s t test. **P < 0.01.