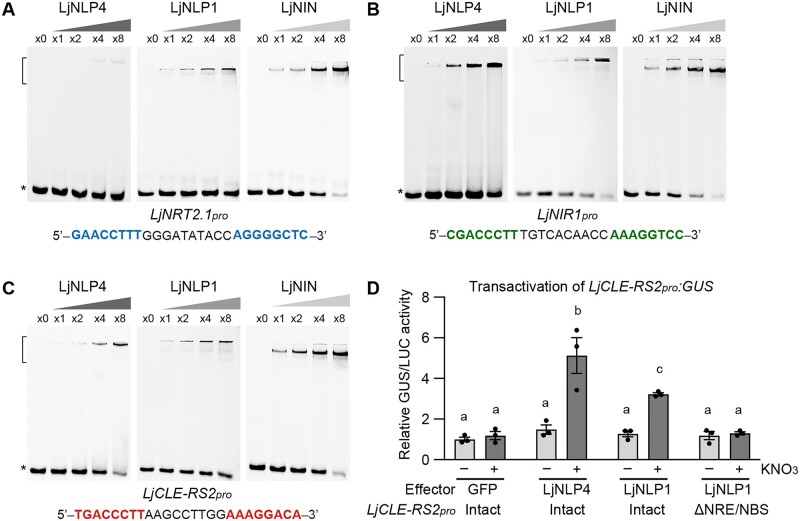
Figure 5.
Protein–DNA interaction of LjNLP4, LjNLP1, and LjNIN. A–C, EMSA showing LjNLP4, LjNLP1, or LjNIN binding to the cis-elements on the LjNRT2.1pro (A), LjNIR1pro (B), and LjCLE-RS2pro (C). MBP-LjNLP4 (564–976), MBP-LjNLP1 (573‒903), or MBP-LjNIN (551–878) recombinant proteins, consisting of an RWP-RK and a PB1 domain, were incubated with the FAM-labeled DNA probe (Supplemental Data Set 1). Blue, green, and red nucleotides indicate conserved motifs among the DNA fragments. Brackets and asterisks, respectively, indicate the position of shifted bands showing protein–DNA interaction and of free probes that did not interact with proteins. D, Transactivation of LjCLE-RS2pro:GUS in L. japonicus mesophyll protoplasts transformed with respective constructs (n = 3 independent pools of protoplasts). GFP, LjNLP4, and LjNLP1 used as effectors. Two types of LjCLE-RS2pro:GUS reporter constructs were used, one with an intact LjCLE-RS2 promoter and one with a modified LjCLE-RS2 promoter in which the NRE/NBS (Figure 5C; Nishida et al., 2021) was specifically deleted (ΔNRE/NBS) (Supplemental Figure S6). Transformed protoplasts were incubated with 0 (−) or 10 mM (+) KNO3. GUS activity was measured relative to 35Spro:LUC activity. Transactivation data were normalized to the condition in which GFP was expressed in the absence of KNO3. Error bars indicate sem. Different letters indicate statistically significant differences (P < 0.05, one-way ANOVA followed by multiple comparisons).