FIGURE 3.
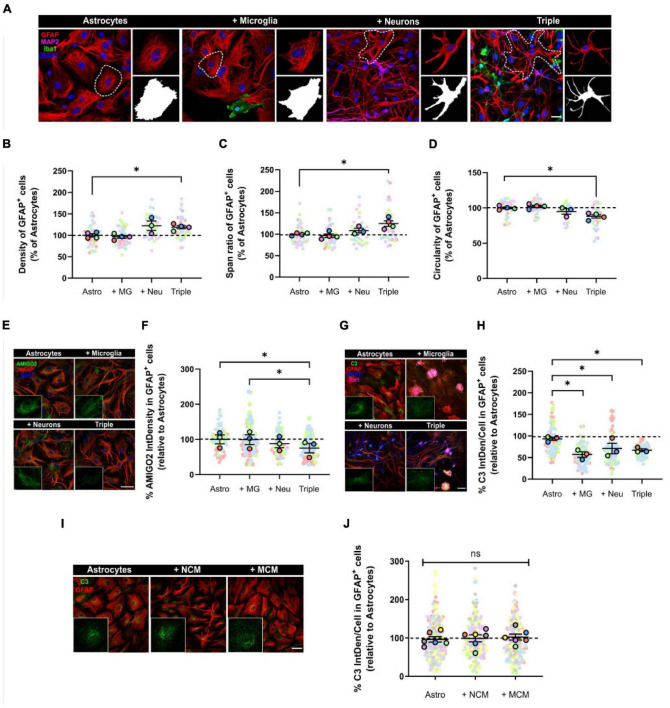
Astrocytes displayed a less reactive morphology and reduced expression of A1 activation markers AMIGO2 and C3. (A) Representative images of the analysis of astrocytic morphology using GFAP and Fractal Analysis in Fiji Software. Black and white images represent the outline of individual cells selected in each condition. Scale bar = 20 μm. (B) Quantification of the density of GFAP+ astrocytes (n = 4). (C) Quantification of the span ratio of GFAP+ astrocytes (n = 4). (D) Quantification of the circularity of GFAP+ astrocytes (n = 4). (E) Representative image of A1 astrocytic marker AMIGO2. Scale bar = 40 μm. (F) Quantification of the expression of AMIGO2 inside GFAP+ cells (n = 3). (G) Representative image of A1 astrocytic marker C3. Scale bar = 40 μm. (H) Quantification of the integrated density of C3 inside GFAP+ cells (n = 3). (I) Representative image of C3 in GFAP+ astrocytes after conditioned media treatment. Scale bar = 40 μm. (J) Quantification of the integrated density of C3 (n = 6). Each bar represents the mean ± SEM. *p < 0.05; ns, not significant. For quantification, 5–6 random fields per condition were used. Neu, neurons; MG, microglia; Astro, astrocytes; NCM, neuron conditioned media; MCM, microglia conditioned media; IntDen, Integrated density.