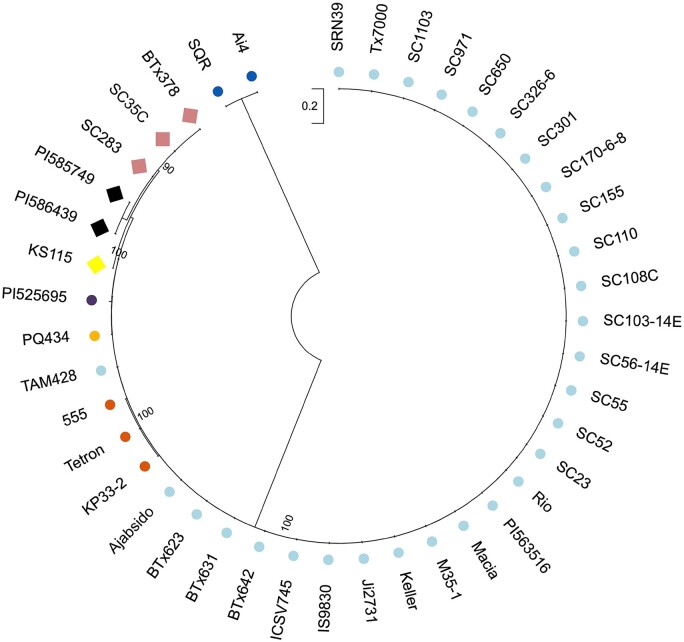
Figure 9.
The maximum likelihood method and JTT matrix-based model analysis of the ARG1 amino acid sequences showing the evolutionary relationship of 42 different sorghum genotypes. Diamonds indicate resistance and circles indicate susceptible genotypes to Cs strain Csgl2. The ARG1 proteins were divided into eight sub-clusters and represented by different colors. Initial tree(s) for the heuristic search were obtained automatically by applying Neighbor-Join and BioNJ algorithms to a matrix of pairwise distances estimated using the JTT model, and then selecting the topology with superior log likelihood value. The tree is drawn to scale, with branch lengths measured in the number of substitutions per site. There were a total of 953 positions in the final data set. Evolutionary analyses were conducted in MEGA X. Bootstrap values are given at the node as a percentage of 1,000 replicates.