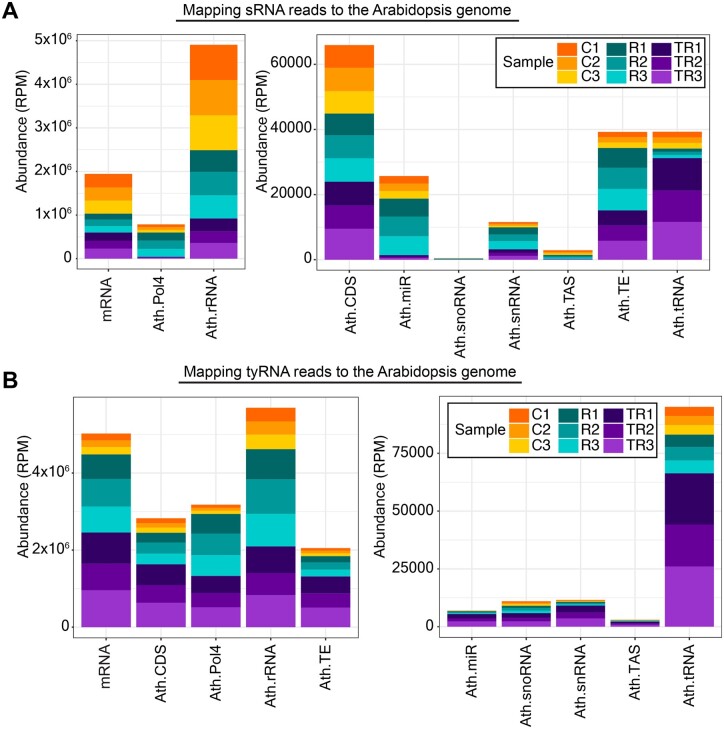
Figure 3.
Apoplastic sRNAs are derived from diverse sources. A, Specific subclasses of sRNAs are protected by proteins. sRNAs that mapped to the genome were categorized by origin and plotted by relative abundance in RPM. Compared to controls (C1–C3), treatment with RNase A alone (R1–R3) increased the relative proportion of Pol IV-, miRNA-, snRNA- and TE-derived sRNAs, whereas treatment with trypsin plus RNase A (TR1–TR3) decreased their relative proportion. This indicated that the majority of these sRNAs are protected by protein and are located outside EVs. snoRNA, small nucleolar RNA; TAS, trans-acting siRNA. B, tyRNAs are mostly located inside EVs. tyRNAs that mapped to the genome were categorized by origin and plotted by relative abundance, as with the sRNAs above. All categories of tyRNAs increased in relative abundance upon treatment with trypsin plus RNase, indicating that they were protected against trypsin plus RNase treatment and hence are mostly located inside EVs. For both panels, the x-axis indicates the RNA source, and the y-axis indicates its abundance in RPM mapped reads. Data from three independent biological replicates (AWF pooled from 24 plants in each replicate) are stacked together in a single bar plot and color-coded as shown in the legend.