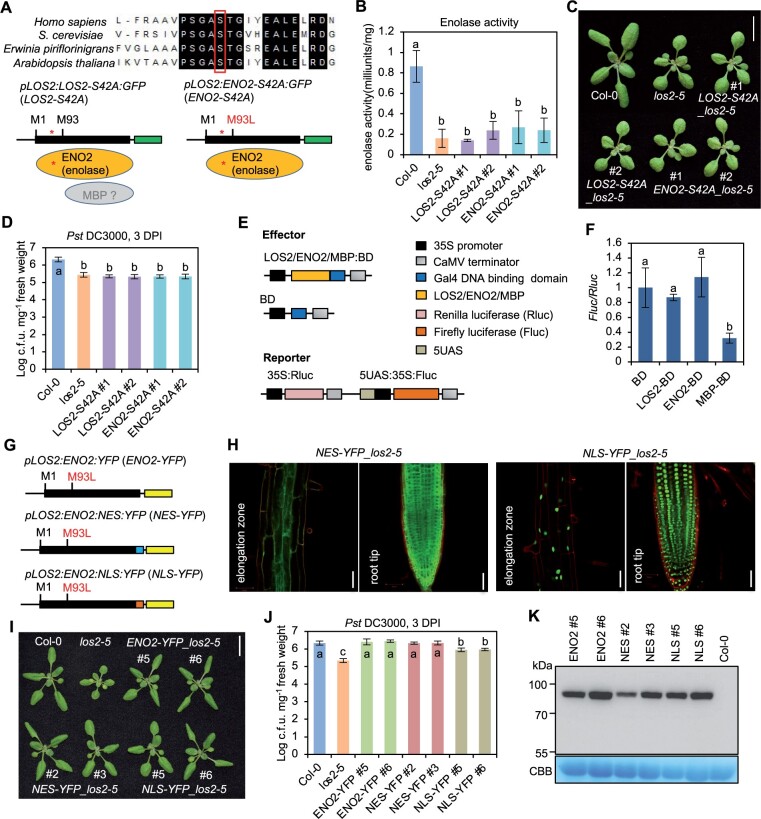
Figure 6.
The enzymatic activity of ENO2 contributes to plant growth and immunity regulation. A, Sequence alignment of enolases from human, yeast, bacterium, and Arabidopsis. The partial alignment result is shown. The red box and star indicate the serine at position 42 in Arabidopsis which was mutated into Alanine (S42A). B–D, Enolase activity (B), morphology (C), and growth of bacterial pathogen Pst DC3000 (D) in Col-0, los2-5 and two representative lines of pLOS2:LOS2-S42A:GFP and pLOS2:ENO2-S42A:GFP in los2-5 background. E, Schematics of constructs used for the dual-luciferase reporter assay. F, Firefly luciferase expression level after normalized to renilla luciferase expression. The expression of BD was set to 1. G, Simplified gene structure for fusion proteins ENO2-YFP, NES-GFP, and NLS-GFP. H, Confocal imaging of fusion proteins in Arabidopsis root treated with propidium iodide. Scale bar = 50 μm. I, Morphology of Col-0, los2-5 and complementation lines of ENO2-YFP (#5 and #6), NES-YFP (#2 and #3) and NLS-YFP (#5 and #6) in los2-5 background. J, Growth of bacterial pathogen Pst DC3000 in Col-0, los2-5, ENO2-YFP (#5 and #6), NES-YFP (#2 and #3) and NLS-YFP (#5 and #6) in los2-5 background. K, Immunoblot detecting the fusion proteins by anti-GFP antibody in complementation lines of ENO2-YFP (#5 and #6), NES-YFP (#2 and #3), and NLS-YFP (#5 and #6) in los2-5 background. Col-0 was used as a negative control. Four and three biological replicates were performed for (B) and (F), respectively. Six biological replicates were performed for (D) and (J). Error bars represent sd. Different letters indicate significant difference tested by one-way ANOVA/Duncan’s multiple range test via R 3.6.3 with “agricolae” package, P < 0.05. Scale bar in (C) and (I) is 1 cm. CBB, Coomassie brilliant blue.