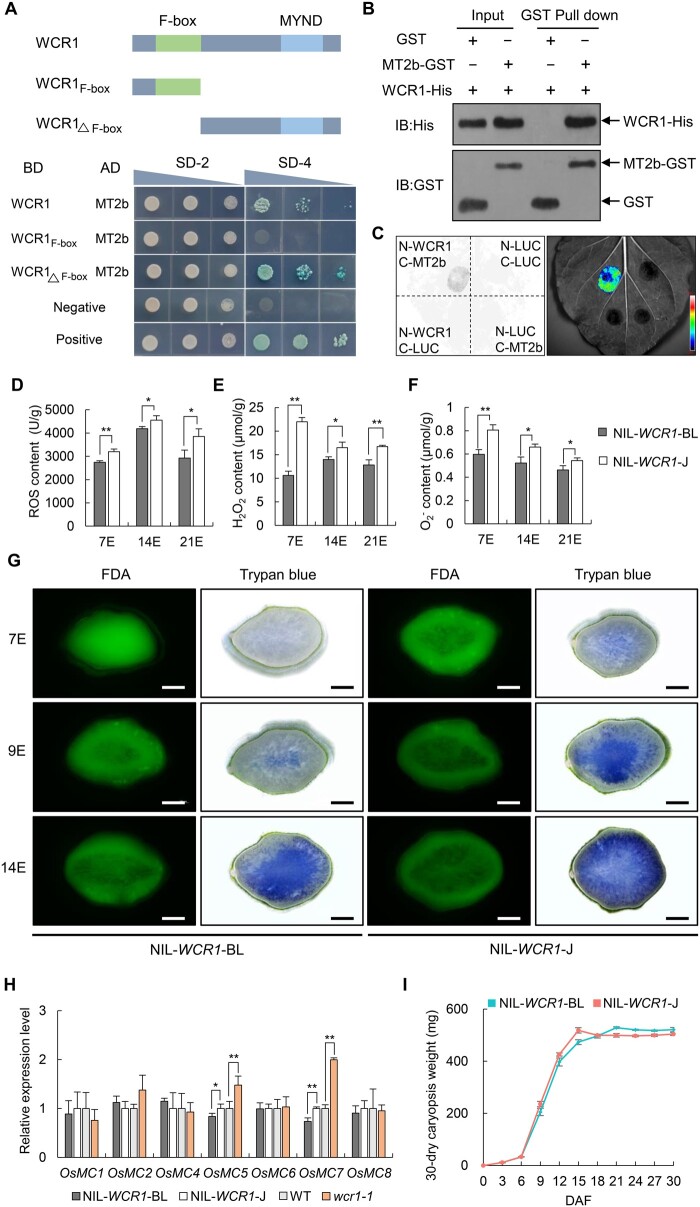
Figure 6.
WCR1 interacts with MT2b to scavenge ROS and delay PCD in developing endosperm. A, WCR1 interacts with MT2b in yeast cells. The N terminus of WCR1 including the F-box domain and the C terminus including the MYND domain are shown. SD-2 and SD-4 represent SD/–Trp–Leu and SD/–Trp–Leu–His–Ade + X-Gal selection medium, respectively. Negative, empty vectors pGBKT7 and pGADT7. Positive, the OsMADS1 gene, which formed a homodimer, was linked to pGBKT7 (BD-OsMADS1) and pGADT7 (AD-OsMADS1), respectively, and yeast harboring these two vectors grew on SD-4 medium. B, Pull-down assay. Proteins were pulled down by GST agarose beads and immunoblotted using His and GST antibodies. C, Split-LUC assay. nLUC-tagged WCR1 and cLUC-tagged MT2b were co-transformed into N. benthamiana leaves. Color bar shows the intensity of LUC signals. D–F, Detection of ROS, H2O2, and contents in endosperm at 7, 14, and 21 DAF (7E, 14E, and 21E). Each endosperm sample was obtained from 30 grains with three repeats. G, Trypan blue and FDA staining of transverse sections of grains at different developmental stages for NIL-WCR1-BL and NIL-WCR1-J. Blue areas and green fluorescence represent areas of dead and living cells, respectively. Scale bars, 500 μm. H, Relative expression levels of PCD-related OsMC genes in 9 DAF endosperm. Endosperm samples were taken from at least four plants. In (D–F and H), significant differences were determined by two-tailed t tests, *P ≤ 0.05, **P ≤ 0.01. Error bars, sem. I, Dynamic changes in mean dry weights of caryopses. Data show the total weights of 30 dry dehulled caryopses with three biological repeats from the upper parts of panicles from different plants. Error bars, sem.