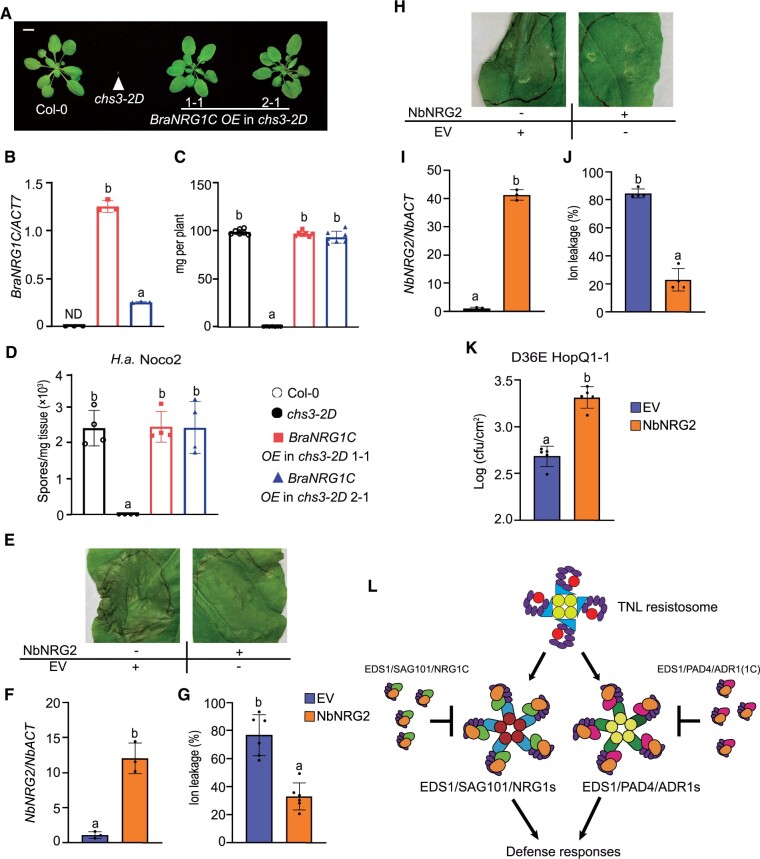
Figure 7.
NRG1C-type N-terminally truncated hNLRs from canola (Brassica napus cv. Westar) and N. benthamiana function similarly to A. thaliana NRG1C. A, Morphology of 4-week-old soil-grown plants of Col-0, chs3-2D, and two independent transgenic lines of BraNRG1C OE in the chs3-2D background. Bar = 1 cm. B, BraNRG1C gene expression in 2-week-old plate-grown plants, as determined by qRT-PCR. Statistical significance is indicated by different letters (P < 0.01). Error bars represent means ± sd (n = 3). ND, not detectable. Three independent experiments were carried out with similar results. The color legends are included in D. C, Fresh weights of the plants in (A). Statistical significance is indicated by different letters (P < 0.01). Error bars represent means ± sd (n = 7). The color legends are included in D. D, Quantification of H.a. Noco2 sporulation in the indicated genotypes at 7 dpi with 105 spores per ml water. Statistical significance is indicated by different letters (P < 0.01). Error bars represent means ± sd (n = 4). Three independent experiments were carried out with similar results. E, HR in N. benthamiana leaves expressing the indicated proteins at 48 hpi following the infiltration of P.s.t. DC3000 D36E HopQ1-1 to deliver HopQ1-1 effector at OD600 = 0.2. The photograph was taken at 4 dpi after P.s.t. DC3000 D36E HopQ1-1 infiltration. F, NbNRG2 gene expression in the infiltrated N. benthamiana leaves indicated in E, as determined by qRT-PCR (EV-infiltrated leaves served as a control whose NbNRG2 transcript level was set at 1.0). Samples were collected at 48 hpi with the indicated proteins. Statistical significance is indicated by different letters (P < 0.01). Error bars represent means ± sd (n = 3). Three independent experiments were carried out with similar results. The color legends are included in G. G, Ion leakage of N. benthamiana leaves upon P.s.t. DC3000 D36E HopQ1-1 infiltration under the same conditions as in E. Statistical significance is indicated by different letters (P < 0.01). Error bars represent means ± sd (n = 6). Three independent experiments were carried out with similar results. H, HR in N. benthamiana leaves expressing the indicated proteins at 48 hpi following the infiltration of Agrobacterium carrying HopQ1-1 effector at OD600 =0.08. The photographs were taken at 2 dpi after Agrobacterium infiltration. Three independent experiments were carried out with similar results. I, NbNRG2 gene expression in the infiltrated N. benthamiana leaves indicated in (H), as determined by qRT-PCR (EV infiltrated leaves served as a control whose NbNRG2 transcript level was set at 1.0). Samples were collected at 48 hpi with the indicated proteins. Statistical significance is indicated by different letters (P < 0.01). Error bars represent means ± sd (n = 3). Three independent experiments were carried out with similar results. The color legends are included in K. J, Ion leakage of N. benthamiana leaves upon Agrobacterium infiltration under the same conditions as in H. Statistical significance is indicated by different letters (P < 0.01). Error bars represent means ± sd (n = 4). The color legends are included in K. K, Growth of P.s.t. DC3000 D36E HopQ1-1 in N. benthamiana leaves expressing EV-3FLAG or NbNRG2 for 48 h. After 48 h, the leaves were infiltrated with P.s.t. DC3000 D36E HopQ1-1 at OD600 = 0.005 and bacterial growth was counted at 4 dpi. Statistical significance is indicated by different letters (P < 0.01). Error bars represent means ± sd (n = 5). Three independent experiments were carried out with similar results. L, A model depicting the negative regulation of NRG1C downstream of TNL activation in Arabidopsis. An effector-induced TNL resistosome with NADase activity (Horsefield et al., 2019; Wan et al., 2019; Ma et al., 2020; Martin et al., 2020; Tian and Li, 2020) signals through two parallel complexes, EDS1–PAD4–ADR1s and EDS1–SAG101–NRG1A/1B, to activate defense responses (Sun et al., 2021; Wu et al., 2021). To avoid over-activation, N-terminally truncated hNLR NRG1C antagonizes immunity mediated by its full-length neighbors NRG1A and NRG1B, while an engineered truncated ADR1 with a similar length to NRG1C [ADR1(1C)] is able to interfere with EDS1–PAD4 dimer to dampen immunity mediated by ADR1s (Wu et al., 2021). Arrows indicate activation, while the “T’” signs represent suppression.