Fig. 1. Pipeline and workflow of this project.
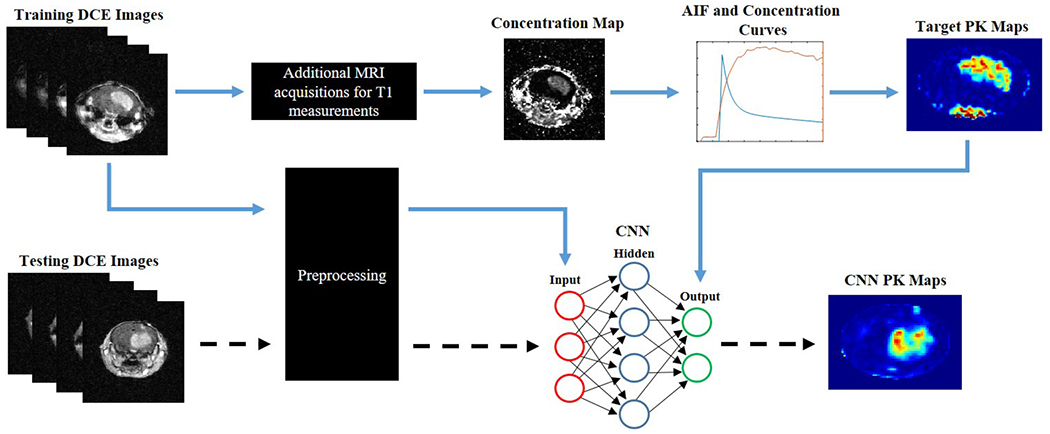
Conventional PK modeling is applied to DCE dynamic images to generate target PK maps. Source DCE dynamic images and target PK maps are fed into the 24-layer CNN for training (blue arrows). An independent testing DCE dynamic imaging dataset can be fed directly into the CNN to generate CNN PK maps following training of the neural network (dashed arrows). Preprocessing of the DCE dynamic images involved reshaping, segmentation, and batch normalization. The neural network can directly predict the PK maps with the DCE time series alone. In contrast, conventional PK modeling requires manual CA arrival identification, additional MRI acquisitions for T1 measurements, complex curve-fitting, as well as the knowledge of the AIF along with the DCE time series in order to estimate these kinetic parameters.
