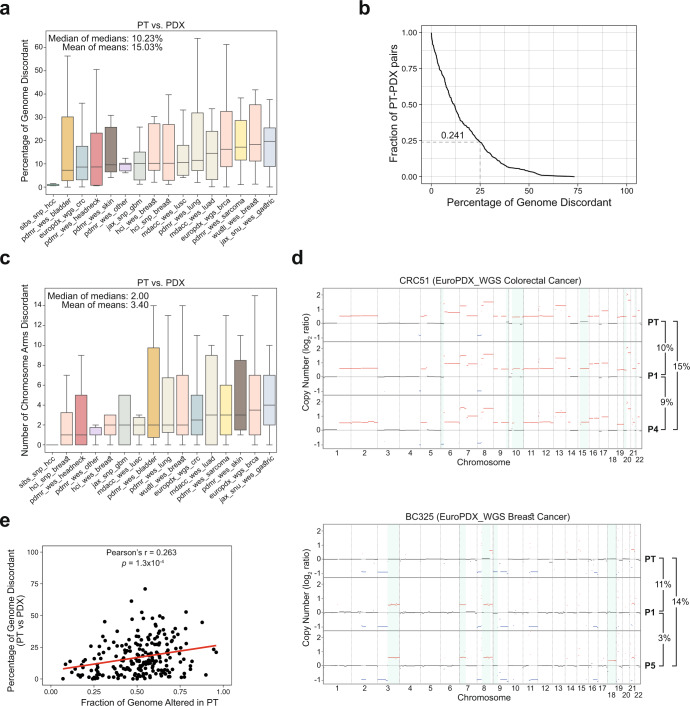
Fig. 1. Thresholds-based comparison of the copy number landscapes of PTs and PDXs.
a A cross-cohort comparison of the percent of the genome that is discordant between matched PT-PDX samples. In the median cohort, a median of 10.23% of the genome is altered between PTs and PDXs. Bar, median; colored rectangle, 25th to 75th percentile; whiskers, Q1 – 1.5*IQR to Q3 + 1.5*IQR; outliers were excluded from the plot. b A reverse estimator of cumulative distribution function (1 - eCDF) plot showing the fraction of PT-PDX pairs in which over a given percentage of the genome is discordant. Over 25% of the genome was discordant in 24.1% of the matched PT-PDX samples. c A cross-cohort comparison of the number of chromosome arms that are discordant between matched PT-PDX samples. A median of 2 chromosome arms are altered between PTs and PDXs across cohorts. Bar, median; colored rectangle, 25th to 75th percentile; whiskers, Q1 – 1.5*IQR to Q3 + 1.5*IQR; outliers were excluded from the plot. d Examples of CN differences between matched PT, an earlier-passage (P1) PDX, and later-passage (P4, P5) PDX samples from the EuroPDX_WGS colorectal and breast cancer cohorts. Red, CN gain; blue, CN loss. Prominent differences are highlighted with a light blue background. The fraction of the genome that is altered between samples is shown to the right of the plot. e A scatter plot presenting the correlation between the fraction of the genome affected by CN in the PT and the discordance between that PT and its derived PDX. In general, the more affected by CN a tumor is, the more divergent its PDX is (Pearson’s r=0.26; p=1.3 × 10-4).