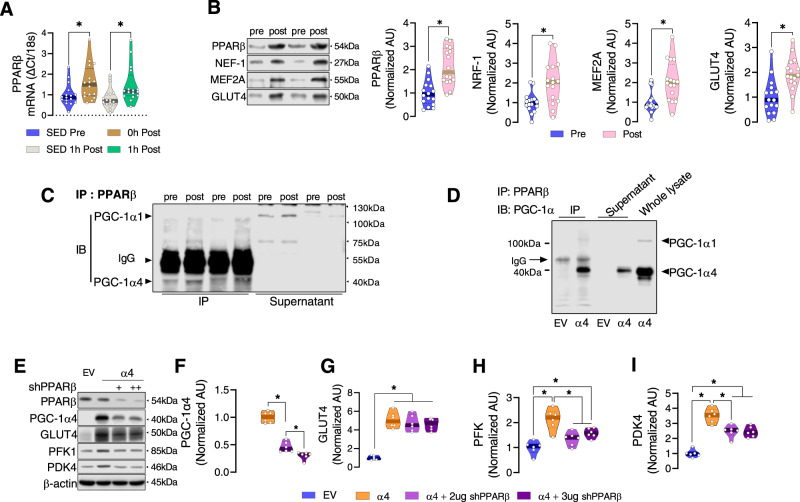
Fig. 5. Resistance exercise training-induced PGC-1ɑ4 cooperates with PPARβ to regulate glycolysis.
A PPARβ mRNA was determined by qPCR in muscle biopsy samples of healthy participants before (SED Pre), within 10 min after (0 h Post), and 1 h after (1 h Post) a one-legged resistance exercise (RE) bout (n = 16). Time-matched control muscle samples were obtained from the non-exercised leg at 1 h post-RE (SED 1 h Post) (n = 16 per group). One-way ANOVA was used with multiple comparisons. B Protein abundance of PPARβ and its downstream related proteins was determined in muscle before (Pre) and after (Post) RET. Representative immunoblots for each protein and quantification of the relative change after training for PPARβ, NRF-1, MEF2A, and GLUT4 are displayed (n = 16). Paired two-tailed t-tests were used. C PPARβ was immunoprecipitated from pooled muscle samples before (pre) and after (post) RET, then the immunoprecipitate was immunoblotted for PGC-1α1 and PGC-1α4 to determine binding between PPARβ with PGC-1α1 and PGC-1α4. Owing to the large amount of protein required to immunoprecipitate enough PPARβ for immunoblot detection of PGC-1α1 and PGC-1α4, samples of multiple participants before and after training were pooled together, resulting in a smaller n for PGC-1α isoform quantification (n = 2). D The binding between PPARβ and PGC-1α isoforms was confirmed using cells overexpressing PGC-1α4. Experiment shown in panel D was performed once. E–I PPARβ was silenced by shPPARβ overexpression in myotubes, then PGC-1α4 was overexpressed (n = 6). The abundance of some of the important glycolytic proteins (GLUT4, PFK1, and PDK4) was determined by immunoblotting. One-way ANOVAs were used with multiple comparisons. *P < 0.05. Values are expressed as individual data points. Significant labeled P-values in each panel from left to right are as follows: panel A = 0.024 and 0.020; panel B = < 0.001, 0.002, <0.001, <0.001; panel F = < 0.001 and 0.010; panel G = < 0.001; panel H = < 0.001, <0.001, and <0.018; panel I = < 0.001, <0.001, and 0.002.