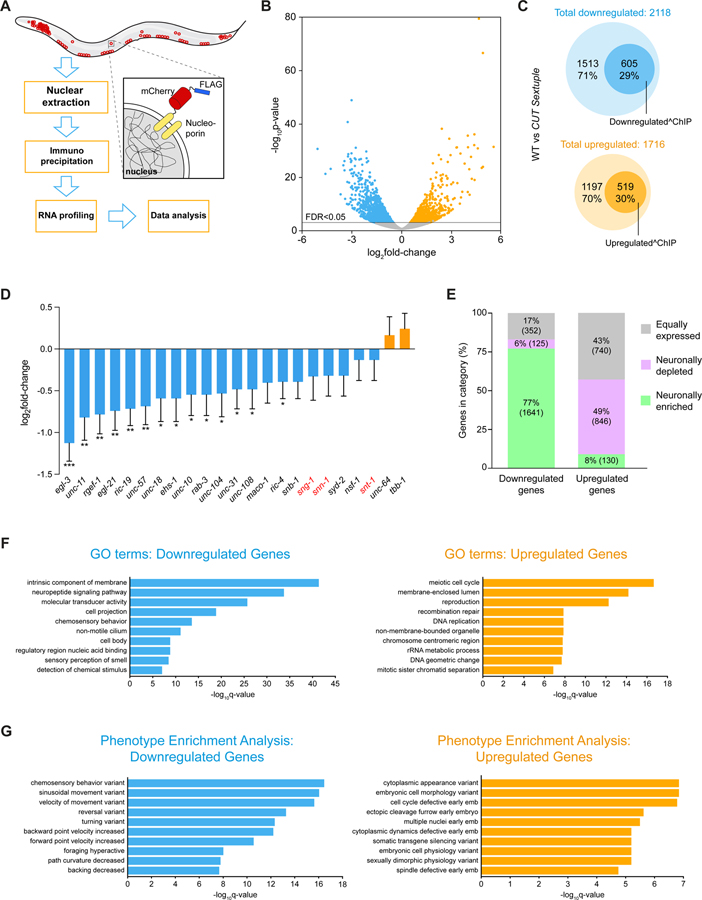
Figure 5. Transcriptional profiling of CUT sextuple mutants.
(A) Schematic and experimental design for INTACT sample collection, protocol, and data analysis for neuronal transcriptome profiling.
(B) Volcano plot of differentially expressed genes in CUT sextuple mutant neurons showing significantly (FDR < 0.05) downregulated (blue) or upregulated (orange) genes (RNA-seq, n = 3). See Data S2A for full list of differentially expressed genes.
(C) Diagrams showing overlap between differentially expressed genes in CUT sextuple mutant and genes bound by CEH-48 or CEH-38 in a wild-type ChIP-seq 17. Downregulated genes are marked in blue, upregulated genes are marked in orange, and the genes that contain CUT peaks are marked with dark circles within both clusters. See Figure S5 for effect on ubiquitously expressed genes containing CUT peaks.
(D) Changes of previously described pan-neuronal gene battery 3 in CUT sextuple mutant animals. The data are presented as the log2FoldChange ± standard error calculated by DESeq2, comparing neuronal samples from wild-type and CUT sextuple mutant. The two-stage step-up method of Benjamini, Krieger and Yekutieli (FDR 10%) was used to calculate the q-values for this subset of genes, analyzing the individual p-values obtained from the DESeq2 comparison. *Q < 0.05, **Q < 0.01, ***Q < 0.001 (RNA-seq, n = 3).
(E) Vertical slices representation of the distribution (in percentage) of the downregulated and the upregulated gene sets between the neuronally enriched (green), neuronally depleted (purple) and equally expressed (gray) gene sets. See Data S3A–C for full list of neuronally enriched and depleted genes. See Figure S6 for the validation of pan-neuronal expression of a neuronally-enriched CUT gene target.
(F-G) GO enrichment analysis (F) and phenotype enrichment analysis (G) using gene sets of significantly downregulated (blue) or upregulated (orange) transcripts. Graphs illustrate the 10 most significant terms. Analysis performed using the Gene Set Enrichment Analysis Tool from Wormbase. See Data S4A–D for full list of enriched terms.
WT, wild-type.