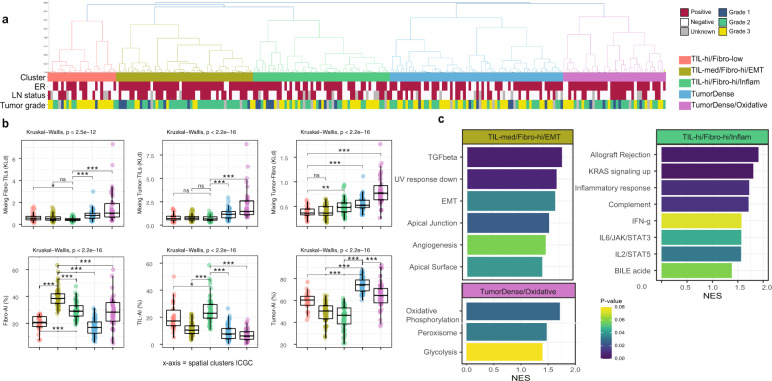
Fig. 3. Hierarchical spatial clustering of ICGC breast tumors results in biological distinct groups.
a Dendrogram of the hierarchical clustering of the ICGC cohort (N = 235) with annotated clinical parameters. b Boxplots of the six spatial H&E measurements that were used for the clustering. Box plots show median, lower, and upper hinges correspond to the 25th and 75th percentile. Y-axis shows KLd (three upper graphs) and percentages (three lower graphs). Kruskal–Wallis test and Wilcoxon-rank test for comparing the highest infiltrated or most mixed cluster to the rest. *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001; NS not significant by Wilcoxon-rank test. c The normalized enrichment score (NES) for the hallmarks of cancer gene sets for each of the five spatial clusters (ER+HER2− samples only, N = 179).