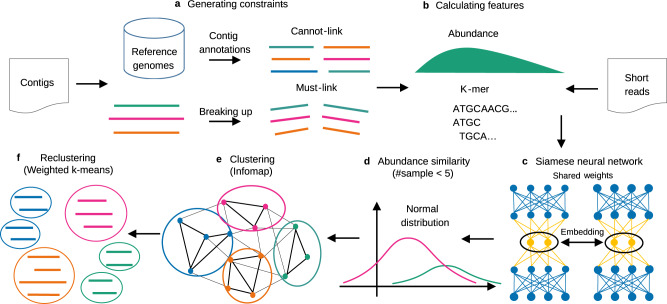
Fig. 1. Overview of the SemiBin pipeline.
a generate must-link constraints by breaking up contigs artificially and cannot-link constraints based on contig taxonomic annotations (i.e. GTDB reference genomes). b calculate abundance estimates (average and variance of the number of reads per base) and k-mer frequency of every contig. c train siamese neural network using the cannot-link and must-link constraints as inputs (k-mer frequencies and abundance features). The learned embedding will be the features from the output layer of the neural network and be used in step e for binning. d Based on the assumption that the number of reads per base obeys a normal distribution, calculate the Kullback-Leibler divergence of the normal distributions of two contigs. SemiBin uses this value as the abundance similarity when the number of samples used is smaller than 5. e generate a sparse graph with the embedded distance and abundance similarity between contigs and uses the Infomap algorithm to obtain the preliminary bins. f SemiBin uses weighted k-means to recluster contigs in the bins whose mean number of single-copy genes is greater than one to get the final bins.