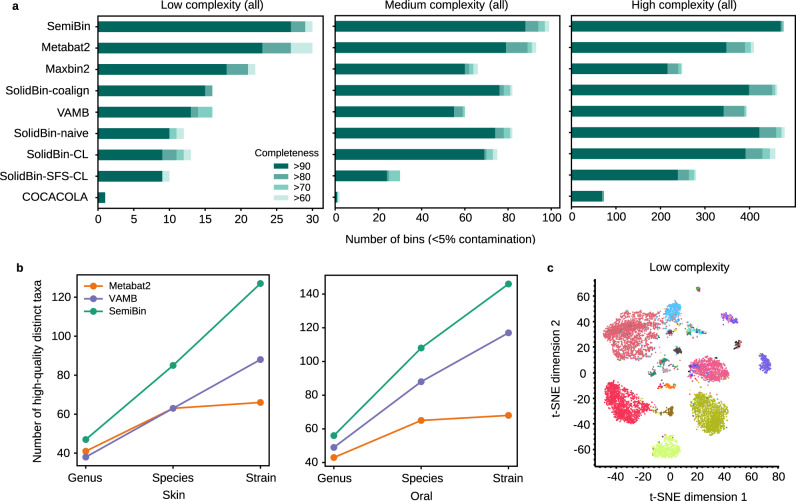
Fig. 2. SemiBin outperformed other binners in simulated datasets with single-sample, co-assembly, and multi-sample binning.
a In CAMI I simulated datasets, SemiBin returned more high-quality bins. Shown are the numbers of reconstructed genomes per method with varying completeness and contamination <5% (methods shown, top to bottom: SemiBin, Metabat2, Maxbin2, SolidBin-coalign, VAMB, SolidBin-naive, SolidBin-CL, SolidBin-SFS-CL, and COCACOLA). b SemiBin reconstructed a larger number of distinct high-quality genera, species, and strains in the CAMI II Skin and Oral datasets compared to either Metabat2 or VAMB. A high-quality strain is considered to have been reconstructed if any bin contains the strain with completeness >90% and contamination <5% (see Methods). If at least one high-quality strain is reconstructed for a particular genus or species, then those are considered to have been reconstructed. c Semi-supervised embedding separates contigs from different genomes. Shown is a two-dimensional visualization of embedding of the low complexity dataset from CAMI I, with contigs colored by their original genome (using t-SNE, as implemented in scikit-learn, parameters: perplexity = 50, init = "pca"77). Source data are provided as a Source Data file.