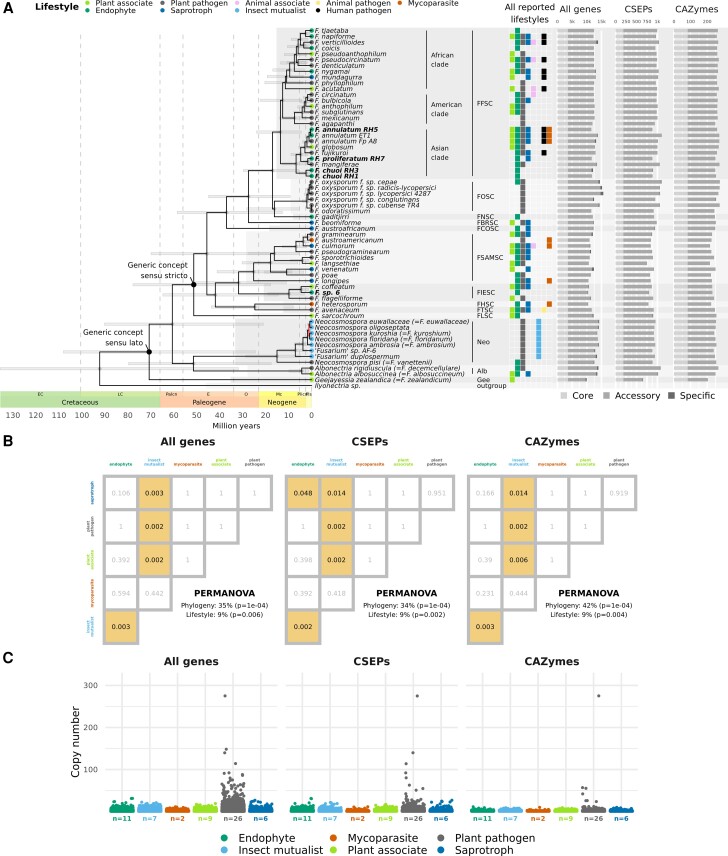
Fig. 2.
(A) Genome-scale phylogeny of fusarioid taxa produced by RAxML-NG from 1,060 core single-copy genes. All branches were significantly supported (≥70 FBP), except those in red. A time scale for node ages estimated by the IR relaxed clock model is shown below the phylogeny, with highest posterior density 95% confidence intervals shown as bars on nodes. For the AR model results and the exact ages and confidence intervals estimated for every node, see supplementary figure S3, Supplementary Material online. Clades corresponding to species complexes (SC) and allied genera are highlighted with alternating boxes and annotated to the right of taxon names (FFSC = Fusarium fujikuroi species complex, FOSC = Fusarium oxysporum species complex, FNSC = Fusarium nisikadoi species complex, FBRSC = Fusarium burgessii species complex, FCOSC = Fusarium concolor species complex, FSAMSC = Fusarium sambucinum species complex, FIESC = Fusarium incarnatum-equiseti species complex, FHSC = Fusarium heterosporum species complex, FTSC = Fusarium tricinctum species complex, FLSC = Fusarium lateritium species complex, Neo = Neocosmospora, Alb = Albonectria, Gee = Geejayessia). Lifestyles of the strains used in this study are indicated by coloured circles on tips, with other lifestyles reported from the literature summarised in the central grid (see supplementary table S1, Supplementary Material online for references). Bar graphs on the right indicate the number of genes, CSEPs and CAZymes for each taxon, with lightest to darkest colour indicating whether genes are core, accessory, or strain-specific. (B) Matrix of P-values showing whether gene, CSEP, and CAZyme content were significantly different between lifestyles according to pairwise PERMANOVA. Coloured boxes indicate significant P values (<0.05). Global PERMANOVA results are reported in the bottom right of plots (see also supplementary table S4, Supplementary Material online). (C) Scatterplot showing variation in gene copy number across all genes, CSEPs and CAZymes for different lifestyles. Points are jittered to reduce overlap. Sample size (the number of strains) is reported under x-axis labels.