Figure 2.
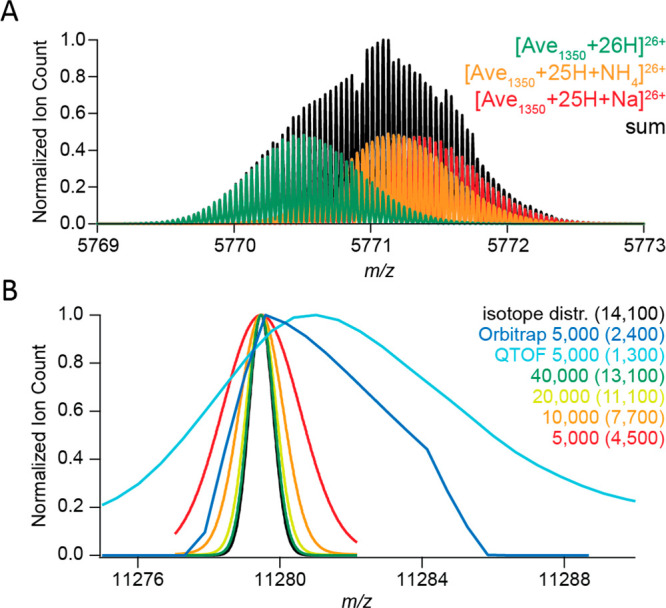
Challenges for resolving isotopologues with high-resolution native MS. (A) Adduct ions affect the mass resolving power. Baseline isotope mass resolution does not permit bare, sodium-bound, and ammonium-bound ions of a 150 kDa averagine protein to be distinguished using native ESI-MS. According to the empirical charging behavior of globular proteins in native MS, 26+ is the most abundant charge state for a molecular weight of 150 kDa. Therefore, the peaks of 26-fold-charged cations of a 150 kDa averagine protein (1350 averagine residues) were generated with MassLynx ver. 4.1, assuming baseline isotope mass resolution (R = 500 000). The isotope distributions of unmodified (green), ammonium-bound (orange), and sodium-bound (red) protein ions were simulated individually and subsequently summed to produce their combined mass spectrum (black). (B) Experimental peaks of globular protein complexes are substantially broader than simulated peaks of their molecular ions. The apparent mass resolution depends on the preset instrumental resolution and the efficiency of adduct removal. Shown are mass spectra of a GroEL ion with a charge of 71+. These were measured on Orbitrap Exactive Plus (blue) and a QToF (cyan) instruments, both operating at an instrument mass resolution of 5000, or simulated with MassLynx ver. 4.1 at mass resolutions of 5000 (red), 10 000 (orange), 20 000 (yellow), and 40 000 (green). The black curve represents the natural isotope envelope of GroEL. Numbers in parentheses correspond to the apparent mass resolutions Rnat determined by measuring the experimental peak widths. Reproduced from ref (32). Copyright 2014 American Society for Mass Spectrometry.
