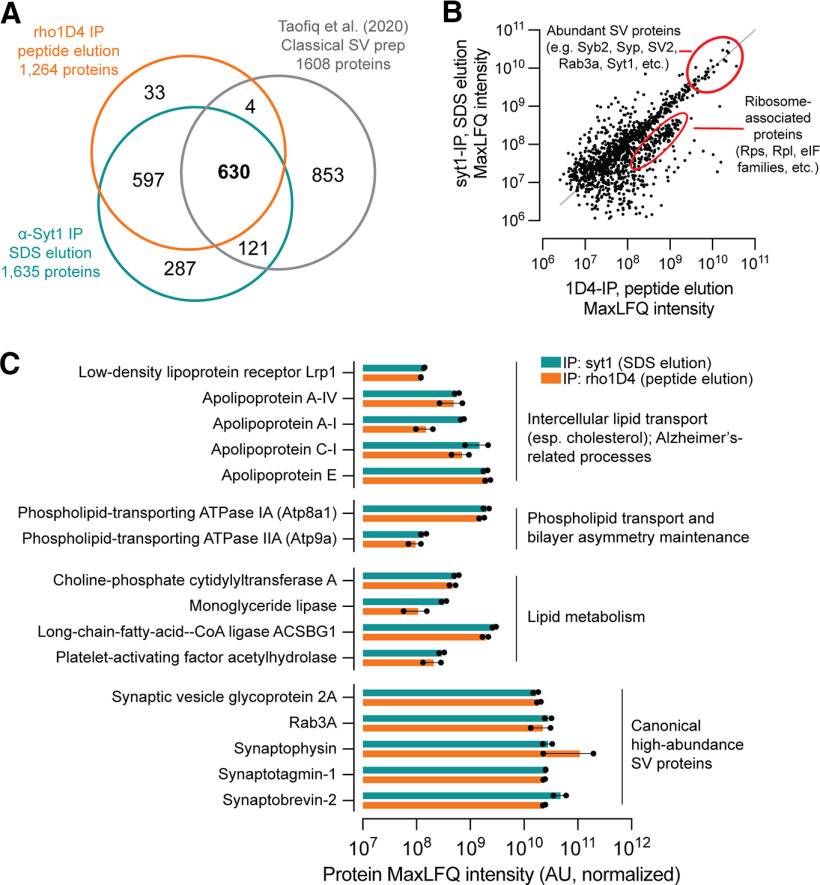
Figure 5.
Synaptic vesicles copurify with proteins involved in lipid homeostasis, including ApoE and the lipoprotein receptor Lrp1. Vesicles were prepared by immunoprecipitation and elution with the 1D4 peptide (rho1D4 mAb beads) or SDS (α-syt1 mAb beads) and subjected to Orbitrap LC-MS (n = 2 biological replicates per purification method). A, Comparison of protein identifications among syt1-IP, rho1D4-IP with peptide elution, and SVs prepared by classical methods (Taoufiq et al., 2020). B, Scatter plot demonstrating robust correlation of intensity values for SVs purified by syt1-IP versus rho1D4-IP with peptide elution. All proteins with quantifiable intensity in at least one replicate were included. A small cluster of proteins enriched in Rho1D4-IP with peptide elution consists of ribosomal proteins that copurify with this method. C, Proteins with well-established roles in lipid transport and metabolism were manually identified from the top 500 most intense proteins in each condition. Closed dots represent values from individual replicates. ApoE predominated among the detected apolipoproteins and was among the top 80 most intense proteins in all replicates, regardless of purification method. Atp9a and Atp8a1 are phospholipid flippases that maintain physiologic distributions of phosphatidylserine. Pcyt1a regulates phosphatidylcholine abundance via synthesis of phosphocholine, a rate-limiting step in phosphatidylcholine synthesis. Monoglyceride lipase cleaves monoacylglycerol molecules and plays key roles in endocannabinoid signaling. The fatty-acid-CoA ligase Acsbg1 activates long- and very-long-chain fatty acids with coenzyme A for downstream metabolic processing. Pla2g7 is a phospholipase A2 isoform important for immune function and breakdown of oxidized lipids. Abundance values for canonical high-abundance SV proteins are given for comparison. Source data used to generate this figure are shown in Extended Data Figure 5-1, and a comparison of proteins detected exclusively by classical approaches (Taoufiq et al., 2020) or by immunopurification (this study) is shown in Extended Data Figure 5-2.