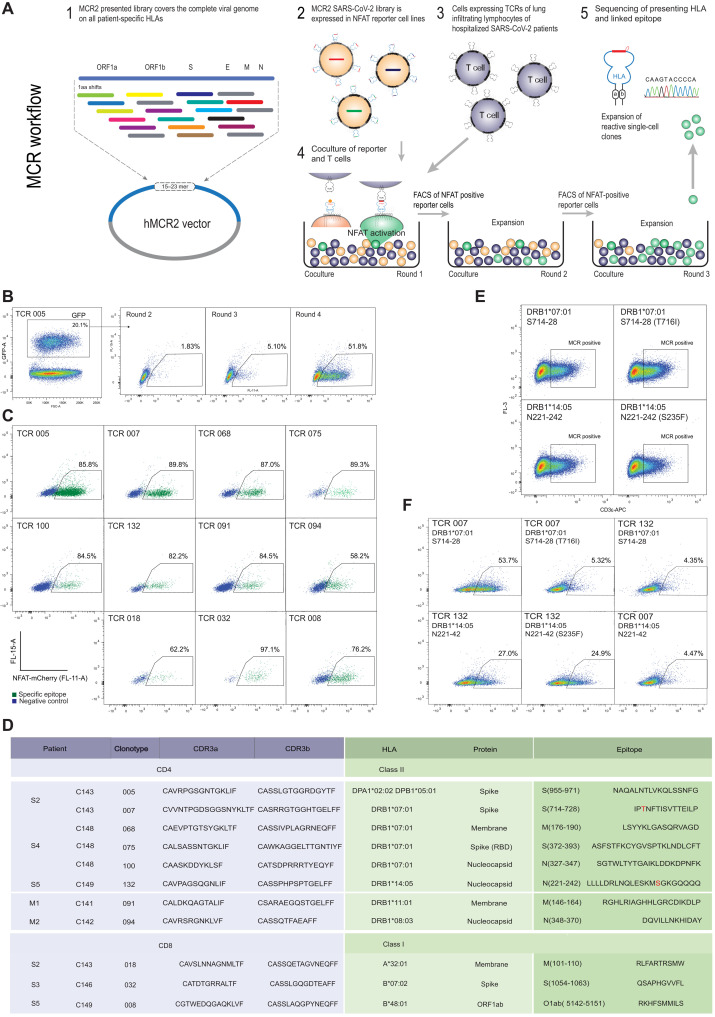
Fig. 4. Deorphaning TCRs from the BAL of patients with COVID-19 by MCR2 screening.
(A) Schematic representation of the TCR deorphaning process. (B) MCR2–SARS-CoV-2+ or SCTz–SARS-CoV-2+ 16.2X reporter cells [green fluorescent protein positive (GFP+)], carrying all possible SARS-CoV-2–derived peptides in the context of all 12 patient-specific HLA alleles (complexity up to 120,000 individual pMHC combinations) were cocultured with 16.2A2 cells transduced with individual TCRs from patients. Responding (NFAT+) reporter cells were sorted, expanded, and cocultured four to five times. FSC-A, forward scatter area. (C) Individual responding reporter clones were isolated and reanalyzed by an additional coculture. (D) Sequences of the deorphaned TCR chains, specific peptides, and HLA restriction. (E) 16.2X reporter cells carrying the MCR2-S714–728 or MCR2-N221–242 were analyzed by flow cytometry for MCR2 expression (by aCD3 staining). (F) 16.2X reporter cells carrying the MCR2-S714–728 or MCR2-S714–728(F716I) (top) and MCR2-N221–242 or MCR2-N221–242(S235F) (bottom) were cocultured with 16.2A2 cells transduced with TCR007 or TCR132, respectively, and NFAT activation was measured on FACS.