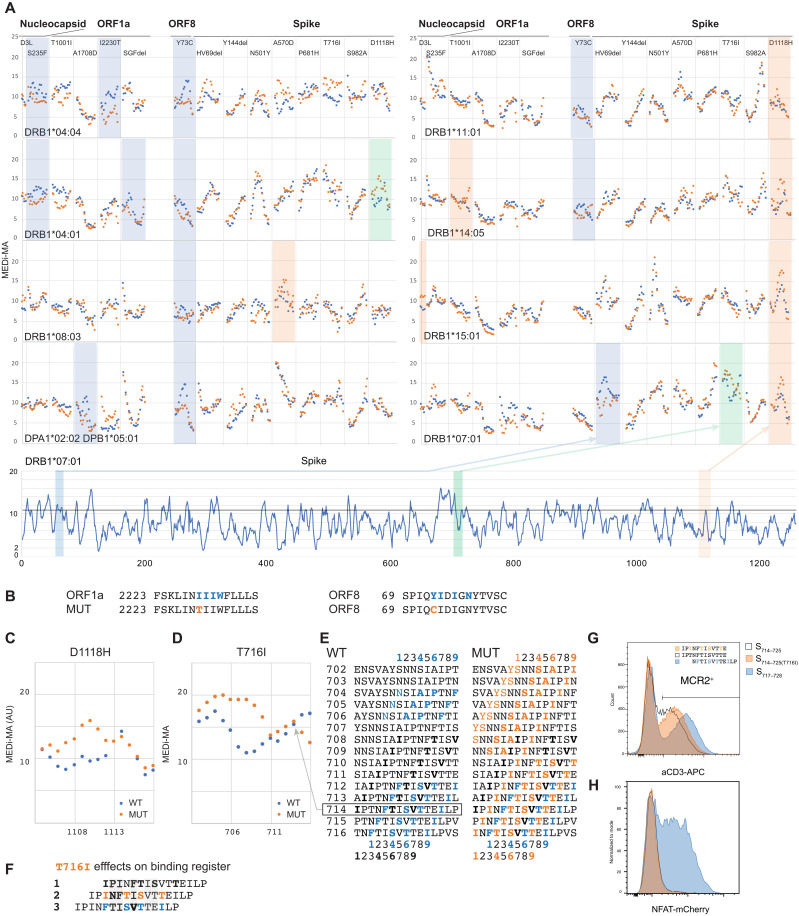
Fig. 6. MEDi reveals candidate immune escape mutants.
(A) Micro MCR2 libraries, containing all 15 (15 amino acid long) peptides spanning the individual mutations were cloned for each HLA and transduced into the 16.2X reporter cells. Individual MEDi-MA scores for the WT (blue dots) and mutated (orange dots) peptides are shown. For context, the MEDi scores for the full Spike protein are shown below. Blue, orange, and green shaded squares indicate differences seen in all repeat experiments (n = 2 or 3). (B) Example peptide sequences from ORF8 and ORF1a with indicated starting residues and the MHC-binding motif for DRB1*04:04 are shown. (C) Detailed view of the MEDi-MA scores for the WT and D1118H Spike-mutated peptides in the context of DRB1*04:01. (D) Detailed view of the MEDi-MA scores for the WT and T716I Spike-mutated peptides in the context of DRB1*07:01. AU, arbitrary units. (E) Fifteen-mer peptides spanning the T716I mutation with indicated starting residues and the different DRB1*07:01-binding motifs. (F) S714–728 peptide sequences with indicated different binding registers forced by several DRB1*07:01-binding motifs present in the WT and/or mutated peptide. TCR-facing residues are highlighted in gray. (G) FACS analysis and sorting of reporter cells transduced with DRB1*07:01-MCR2 carrying the 12-mer peptides: S714–725, S714–725(T716I), and S717–728. (H) Reporter cells from (F) were cocultured with 16.2A2 cells transduced with TCR007, and NFAT activation was measured by flow cytometry.