Figure 5.
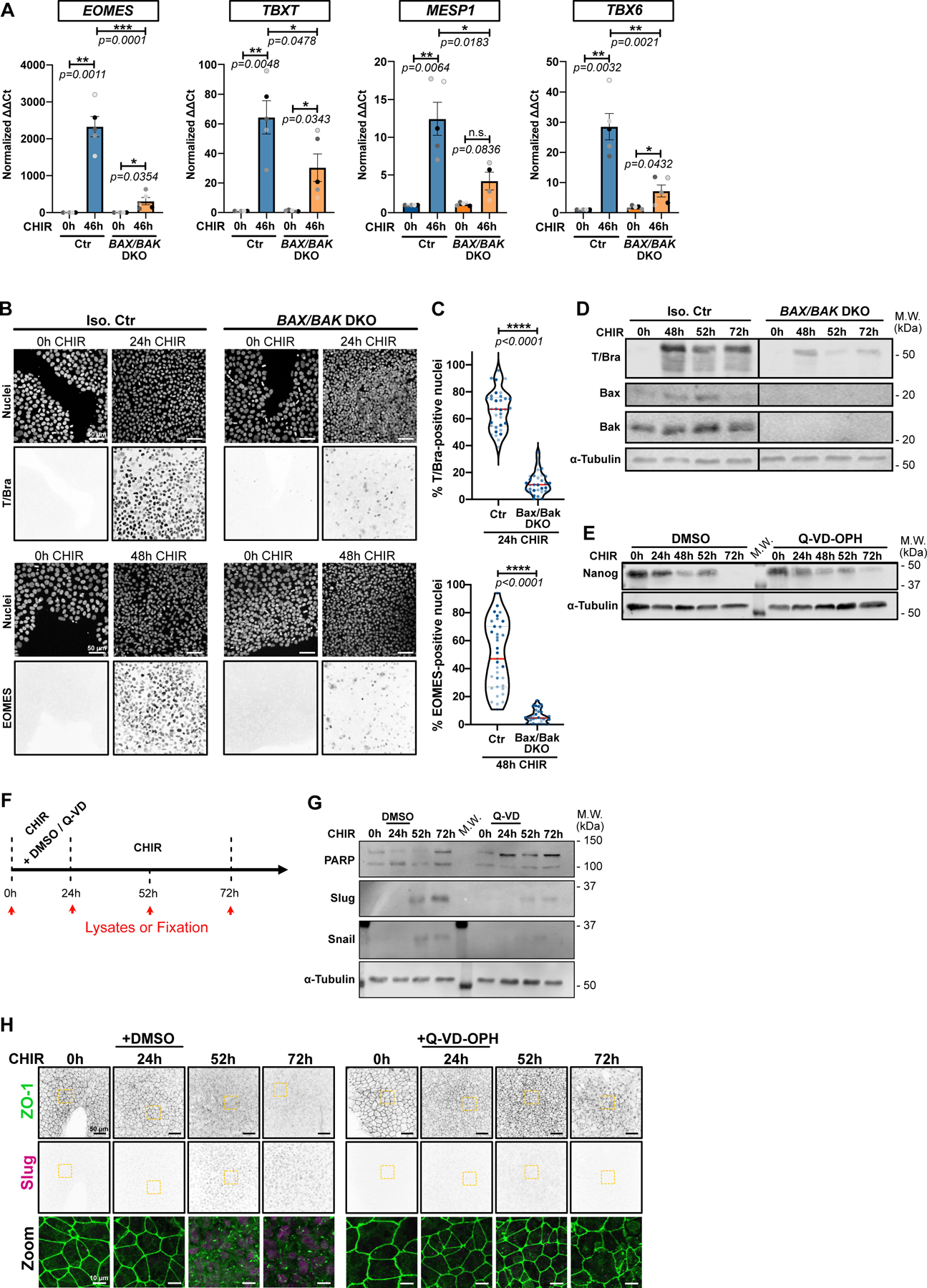
A) Relative gene expression of EOMES, TBXT, MESP1, TBX6 obtained from isogenic control (Ctr) or BAX/BAK DKO hiPSCs, induced or not with CHIR for 46hrs and analyzed by qRT-PCR. ΔΔCt values were normalized to un-induced cells. Biological repeats are color-coded (N=5 independent experiments). Error bar = Mean +/− S.E.M. Two-tailed paired t-test was applied to compare 0hrs vs 46hrs and unpaired t-test was applied to compare Ctr vs DKO.
B-C) Max IPs images of isogenic control (Iso. Ctr) and BAX/BAK DKO hiPSCs stained for T/Bra (top) and EOMES (bottom) and nuclei, before (0hrs) and after CHIR treatment. Scale bar = 50 μm. (B). Percentage of T/Bra-positive (top) and EOMES-positive (bottom) nuclei after CHIR treatment (Median: plain red line – Quartiles: black dotted lines). Biological repeats are color-coded (N=35 random fields of view for EOMES quantification in Iso. Ctr cells and n=34 random fields of view for the remaining data set, all obtained from 3 independent experiments). Mann-Whitney test was applied (C).
D) Immunoblot of isogenic control (Iso. Ctr) and BAX/BAK DKO hiPSCs analyzed for T/Bra expression following CHIR addition.
E) Immunoblot of WT iPSC treated with CHIR +/− 10 μM Q-VD-OPH and analyzed for Nanog expression.
F) Experimental design schematic. Lysate collection and sample fixation time is depicted by red arrows. hiPSCs were treated with CHIR +/− 10 μM Q-VD-OPH for 24hrs, then Q-VD-OPH was washed out and cells were incubated for another 28hrs or 48hrs with CHIR only (respectively 52hrs and 72hrs time point).
G-H) Representative immunoblot (G) and Max IPs images (H) obtained as explained in (F). Membranes were blotted for PARP (cell death marker), and expression level of Snail/Slug (EMT markers) were analyzed. α-Tubulin was used as loading control (G). Scale bar = 50μm. Magnified area (yellow dotted square) is shown as a merge. Scale bar = 15μm.
Source numerical data and unprocessed blots are available in source data.
