Abstract
The present study aimed to synthesise bio-molecule functionalized silver nanoparticles (AgNPs) using leaf extract from mulberry variety S-1635 (Morus alba L.) and to explore its antibacterial efficacy against multidrug resistant (MDR) gut bacteria isolated from natural infection observed from silkworm larvae in rearing conditions. AgNPs formation was established by surface plasmon resonance at 480 nm. The crystallinity of the synthesised AgNPs was checked by HR-TEM and XRD analysis. SEM and TEM characterisation further exhibited the spherical, monodispersed, well scattered nature of the AgNPs with an average particle size of 11.8 nm ± 2.8. The presence of (111), (200), (220) and (311) planes in Bragg's reflections confirmed the face-cantered-cubic crystalline silver. EDX analysis confirmed the presence of elemental silver. FT-IR spectra revealed functional groups were responsible for the reduction of silver ions. The zeta potential value of −17.3 mV and −25.6 mV was recorded in MH and DMEM/F-12 media, respectively. The LC-QTOF/MS and HRMS spectra disclosed the presence of bioactive compounds like flavonoid, gallic acid, and stigmasterol, which are probably involved in the reduction and functionalization of AgNPs. The antibacterial efficacy of bio-molecule functionalized AgNPs and the naked AgNPs was tested on Gram-positive and Gram-negative bacteria isolated from silkworms and characterized by using 16S rDNA and gyrB genes. The cytotoxicity of AgNPs was tested on WRL-68, HEK-293, ACHN, and HUH-7 cell lines using MTT assay. This study provides an insight into the application of bio-molecule functionalized AgNPs for combating various silkworm pathogens which severely affect the agro-rural economy of developing countries.
We aimed to synthesise bio-molecule functionalized silver nanoparticles using leaf extract from mulberry variety S-1635 (Morus alba L.) and to explore its antibacterial effect on multidrug resistant gut bacteria isolated from natural infection observed from silkworm larvae.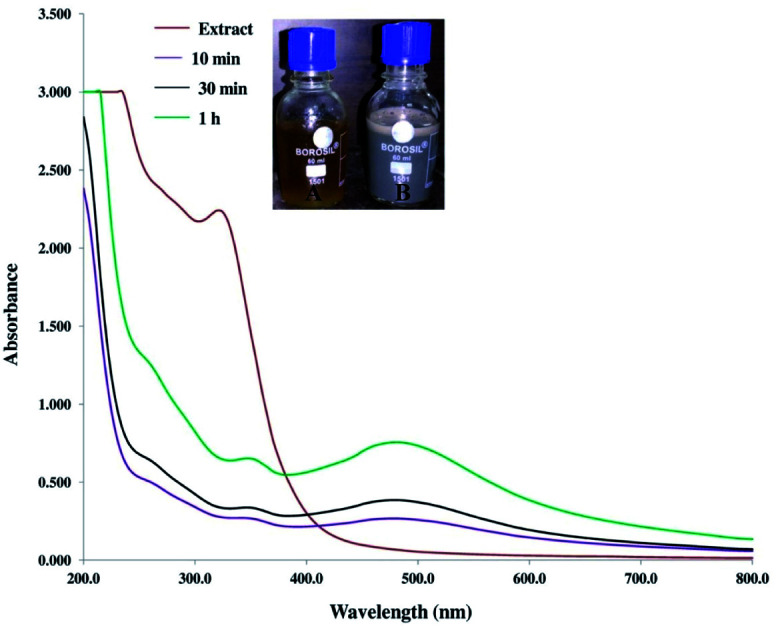
1. Introduction
Nanoscale materials are the principle constituent of nanoscience and nanotechnology. Silver nanoparticles (AgNPs) are used in the various fields of nano-biotechnology.1,2 Noble metallic nanoparticles are used extensively as a therapeutic drug to combat multidrug-resistant (MDR) bacteria.2 These particles have a high surface area per volume proportion, catalytic and optical impact, wide morphological variety, cytotoxic effects, quantum confinement, and electrical and thermal conductivity.3–7 Currently, AgNPs are widely used, ranging from home disinfectants, cosmetics, food packaging industry, textiles, drug delivery, bio-sensor and different medical devices to water purifiers.8–12 Various physical, chemical and biological techniques are used to synthesise AgNPs. The chemical and physical techniques have mainly focused on reducing hazards and difficulties, as synthesis is currently very time consuming and, expensive, and biomedical use of these metallic nanoparticles is hampered by toxicity.13–15 Therefore an eco-friendly, rapid, easy, and less expensive synthesis route is sought for the fabrication of biocompatible AgNPs. This ‘green route’ is characterised by the use of biodegradable materials such as bacteria, fungi, algae, and plant extracts in the biosynthesis of metallic nanoparticles.16–19 Plant extracts are widely used in the NPs synthesis route because of low production cost along with a high reaction rate compared to the microbial synthesis of NPs. Various phytochemicals or bioactive compounds which include flavonoids, terpenoids, polyphenols, and alkaloids, function as reducing agents of Ag+ ions and as stabilisers in the fabrication of biocompatible NPs.20–22 Studies have been done on the use of aqueous leaf extract of Melia azedarach,23 a root extract of Euphorbia wallichii,24 fruit extract of Alpinia nigra,25 and inflorescence extract of Mangifera indica26 to synthesise AgNPs functionalized by bio-molecules.
The mulberry or Morus belongs to the family Moraceae and is described as either a shrub or a small tree. The plant is noted for its economic value as well as its medicinal properties. The M. alba L. variety S-1635 is a triploid variety of mulberry first developed at the Central Sericultural Research & Training Institute (CSRTI), Berhampore, Central Silk Board, Government of India. This variety is widely cultivated to feed the mulberry silkworms engaged in the commercial production of silk and is frequently found in Southern, Eastern, and North-eastern India.27,28 Regarding its uses in medicine and chemistry, various studies have been done. One study showed that the phytochemicals or bio-active compounds in the leaf extract of M. indica L., a member of the genus Morus, exhibit excellent hypolipidemic and hypoglycaemic properties in diabetic patients.29 In another study, Andallu and Varadacharyulu (2003) reported that antioxidant and anti-hyperglycaemic properties were found in the leaf of mulberry plant.30 Earlier study demonstrated the use of aqueous leaf extract of mulberry to synthesise bio-compatible AgNPs.31 Adavallan and Krishnakumar (2014) reported that bio-active compounds in leaf extract of M. alba functioned as bio-reducing and stabilising agents in the biosynthesis of gold nanoparticles (AuNPs).32 All these studies reveal that the leaf extract of mulberry plants shows excellent potential for manufacturing bio-functionalized metallic NPs.
The domesticated silkworm, Bombyx mori L. (Lepidoptera: Bombycidae), an insect of economic importance, is a powerful experimental model for the basic and applied research due to its ease of rearing. Gut microbiome plays a pivotal role in the growth and development of silkworms. Infection during the rearing stages significantly reduces yield, causing substantial financial loss to the agro-based industry.33,34 An artificial diet in silkworms hampers metabolic homeostasis, which in turn has adverse effects on their gross immune system, making them more susceptible to pathogenic attacks.35 Through the eating of contaminated mulberry leaves, the pathogens are transmitted horizontally to silkworm larvae.36 In 1845, there was an epidemic-level outbreak of disease in mulberry silkworms in France, with microbial infections as the major reason.37 Close observation of this debacle pushed Louis Pasteur to pursue his ideas on vaccination against various infectious germs.38 One of the common bacterial diseases among mulberry silkworm is Flacherie, caused by several pathogenic bacteria that belong to a wide variety of genera, which include Aeromonas, Bacillus, Enterobacter, Pseudomonas, Serratia, Streptococcus, and Staphylococcus.39–41 Lu (1991) classified bacterial infections in mulberry silkworm into three groups; toxicosis, bacterial gastroenteric disease and septicemia.42 Cappellozza et al. (2011) were the first to report that Enterococcus mundtii is responsible for Flacherie.43 Silk quality and rate of production are negatively affected by microbial infection in silkworms, as well as by their feeding behaviour. Earlier study revealed that silkworm bed disinfectants such as bundh powder, slaked lime powder and rakshak showed very poor antimicrobial activity against mulberry silkworm pathogens.44 Thus, it is extremely important to find another way to fight these infections using potent antimicrobials. The antimicrobial activity of AgNPs against Gram-positive and Gram-negative bacteria has been confirmed in several other studies: one study showed that, biosynthesised nanosilver showed bactericidal property against Bacillus subtilis, Bacillus vallismortis, and Escherichia coli.16 Ganesh Prabu et al. (2013) reported that AgNPs exhibited excellent antimicrobial action against three strains of the silkworm's bacterial pathogens: Gram-negative bacilli-I &II and Gram-positive cocci.40 To the best of our knowledge this is the first report on the green synthesis of AgNPs using leaf extract of mulberry. The present study focused on the synthesis of AgNPs, the evaluation of antibacterial activity on MDR gut bacterium in cultured silkworm, and on the particles' cytotoxic effect in both normal and cancer cell lines. The synthesised AgNPs have been extensively characterised by UV-vis, FT-IR, HRTEM, EDX, XRD and DLS.
2. Experimental section
2.1. Chemical and biological materials
All the chemicals used in this present study were of analytical grade of which the silver nitrate (AgNO3) (cat. no. 101512) Merck, USA and naked AgNPs (cat. no. 730785) Sigma-Aldrich. Bacterial culture media were procured from HiMedia Laboratories, Mumbai, India. All the solutions were prepared using 0.2 μm filtered autoclaved double-distilled water. The glass ware and labware were baked at 240 °C for 24 h to remove exogenous DNase or RNase, if any. The pipetting devices and counter space were treated with 70% ethanol. The M. alba L. S-1635 plant (specimen no. UD/RG/01) was identified and authenticated from Central National Herbarium, Botanical Survey of India, Government of India, Howrah (ref. no. CNH/Tech. II/2020/7) before the preparation of aqueous leaf extract for AgNPs synthesis.
2.2. Preparation of aqueous leaf extract
A fresh and healthy shoot of the mulberry collected in the Department of Sericulture, Raiganj University, West Bengal, India. Leaf materials (except more or less 2–3 from the top of the shoot) were separated from the stem and placed in a plastic tray. Leaves were washed four times with de-ionised water to remove residual debris and air-dried at ambient temperature for further use. Fifteen grams of finely dissected leaves were boiled with 150 mL sterilised deionised water at 90 °C for 45 min. The extract was then left to cool at ambient temperature and filtered to eliminate any particles through Whatman filter paper no. 41. A pale yellow leaf extract was obtained and stored in the refrigerator (4–8 °C) for further study.
2.3. Biosynthesis of AgNPs
Twenty-five millilitre of leaf extract was transferred to a solution of 50 mL, 10 mM AgNO3 and kept at room temperature with continuous stirring at 250 rpm. The detail step-by-step process was presented in Scheme 1. Successive experiments were performed with same condition. The effect of the Ag+ ion concentration on the formation of AgNPs was checked by varying the concentration of AgNO3 to 1.0, 2.0, 5.0, and 10 mM. The resulting AgNPs were centrifuged to separate them from the supernatant, and the remaining unreacted silver was further washed with deionised water and dried at 50 °C for 24 h and stored for further characterisation.
Scheme 1. Diagrammatic illustration of the biosynthesis of silver nanoparticles using the aqueous leaf extract of mulberry as reducing and functionalizing agents.
2.4. Characterisation
2.4.1. UV-vis spectroscopy
The characterisation of AgNPs was initially executed through UV-visible spectroscopy to inspect the reduction of Ag+ ions to Ag°. The surface plasmon resonance (SPR) absorption spectra of the synthesised AgNPs in the presence of mulberry leaf extract were recorded at varied time intervals in a UV-vis spectrophotometer (Varian Inc., USA) using a quartz cuvette of 1 cm optical path length within the range of 200–800 nm.
2.4.2. X-ray diffraction (XRD) analysis
The crystalline characterisation was conducted in the range of 20° ≤ 2θ ≤ 80° at 40 keV using a Rigaku SmartLab (Japan) X-ray diffractometer, operating at 9 kW and CuKα radiation (λ = 1.54056 Å). The lattice constant of the crystals was designed using PowderX software.45
2.4.3. Microscopic analysis
The surface morphology of biosynthesised AgNPs was studied using a Jeol JSM-6360 scanning electron microscope (Japan) with working distance at 8 mm and accelerating voltage of 30 kV, at SAIF-NEHU, Shillong, India. A drop of sample was placed onto the clean electric stub and allowed to dry. The dried sample was mounted on an aluminium stub with double adhesive tape and was ready for imaging. The shape and size of the AgNPs were measured using high resolution-transmission electron microscopy (HR-TEM) equipped with an energy-dispersive X-ray spectroscopy (EDX) attachment. The samples were carefully prepared by putting a single drop of aqueous biosynthesised AgNPs on a copper-coated grid for analysis. A TEM micrograph was recorded using a Jeol JEM-2100 electron microscope (Japan) worked at the voltage of 200 kV, at SAIF-NEHU, Shillong, India.
2.4.4. Fourier-transform infrared spectroscopy (FT-IR)
FT-IR spectral analysis of aqueous leaf extract and biosynthesised AgNPs was inspected by the FTIR spectrophotometer (Thermo Scientific Nicolet 380) as described previously.46
2.4.5. Dynamic light scattering (DLS)
Dynamic light scattering (Zetasizer Nano ZS90 ZEN3690, Malvern Instruments Ltd., UK) was used to determine the hydrodynamic size (dh), polydispersity index (PDI) and zeta potential of the synthesised bio-molecule functionalized AgNPs as described previously.47
2.5. Liquid chromatography-mass spectrometry (LC-MS)
The aqueous leaf extract of mulberry was characterised using LC-MS/MS to examine the various phytochemical ingredients. The experiment was conducted using a 2D-nanoACQUITY UPLC System furnished with SYNAPT G2 mass spectrometry (Waters, USA). A positive mode of electrospray ionisation (ESI) was also used in this experiment. The source and desolvation temperatures were fixed as 100 °C and 350 °C, respectively. The rates of cone gas flow and desolvation gas flow were set as 50.0 L h−1 and 700 L h−1, respectively. The identification of metabolites in the leaf extract made use of METLIN libraries as well as retention indexes with previously published literature.
2.6. Molecular characterisation of the cultivatable bacteria from silkworm
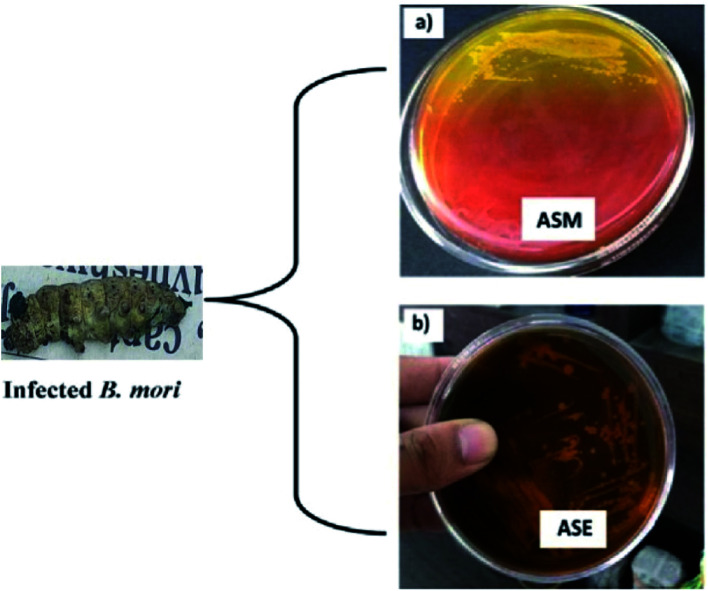
Infected fifth instar larvae of Bombyx mori L. were collected from a silkworm-rearing house located in Kanua village under the Chanchal sub-division of Malda district, West Bengal, India. The larvae were brought to the laboratory in a sterilised container for further study. The larvae were surface sterilised with 5% (w/v) sodium hypochlorite for 5 min after washing it with 70% ethyl alcohol and finally washed three times with sterilised deionised water. After that, a sterilised needle was used to pierce the integument of infected Bombyx mori L. for the withdrawal of body fluid. The body fluid was then transferred to nutrient broth (Himedia, MM244) and kept for incubation at 37 °C for 12 h to allow the growth of culturable bacteria. The inoculum was streaked on mannitol salt agar (Himedia, M118) and eosin methylene blue agar (Himedia, M317) separately for isolation of pure colonies and named as ASM and ASE, respectively. The plates were then incubated overnight at 37 °C, and the most prevalent colony was picked from each plate, based on their colony shape, margin, surface and opacity and maintained on nutrient agar (Himedia, M001) slants at 4 °C for future use with a recommended limit of one week.
2.6.1. Phylogenetic analysis of silkworm infecting bacteria
The isolated strains were freshly grown in 5–6 mL Luria–Bertani broth (Himedia, M1245) at 37 °C with shaking (180 rpm) for 18 hours from the slant cultures. Genomic DNA of isolates was extracted by phenol:chloroform method, as described by Marmur (1961).48 The 16S rDNA was amplified by using universal primer 8F and 1492R according to Turner et al. (1999).49 In addition to 16S rDNA, the house-keeping gene gyrB (DNA gyrase B subunit) for isolate ASE was also amplified by using UP-1 and UP-2r primers, according to Yamamoto and Harayama (1995).50 Purified 16S rDNA and gyrB products were sequenced following the Sanger sequencing method. Both strands of sequenced nucleotides were aligned through BioEdit software (http://www.bioedit.software.informer.com) to retrieve the partial/complete sequence of the 16S rDNA gene of both the isolates and gyrB gene of isolate ASE. The retrieved 16S rDNA sequences of both the isolates were submitted to the EZBioCloud server (https://www.ezbiocloud.net/) to identify the closest type strains. From the EZBioCloud server, all the closest type strains for both the isolated strains were downloaded to obtain multiple sequence alignment with the ClustalW program using MEGA6 software. Phylogenetic trees were constructed by the neighbor-joining (NJ) method, where the evolutionary distances were calculated by the Kimura 2 parameter model.51 The gyrB gene sequence (nucleotide) was compared with BLASTn server (GenBank, NCBI), followed by retrieving the sequences of all the best matches to obtain multiple sequence alignment with the ClustalW program using MEGA6 software. Phylogenetic trees were constructed by the NJ method, where the evolutionary distances were calculated by the Kimura 2 parameter model. Bootstrap analysis for both the phylogenetic trees was executed with resampling 1000 times for the validation of each clade of the phylogenetic tree. The NCBI accession numbers for the 16S rDNA sequences of the selected strain ASM and ASE are MT023669 and MT023436, respectively.
2.7. Antibiotic susceptibility testing
For the antibiotic susceptibility test intended for strain ASE against different antibiotics, a disc diffusion test was performed following standard protocol.52 Antibiotic discs were brought from HiMedia Laboratories. Tests were performed on Mueller–Hinton agar, where freshly grown overnight culture of isolate ASE in Mueller–Hinton broth was spread as inoculum and discs were placed on top of the Mueller–Hinton agar plates. The Petri dishes were then incubated at 37 °C in a BOD incubator overnight. The antibiotics to which strain ASE were sensitive showed halo inhibition zones, whereas the antibiotic discs which were unable to show any halo inhibition zone indicated that strain ASE were resistant.
2.8. Antibacterial property of AgNPs
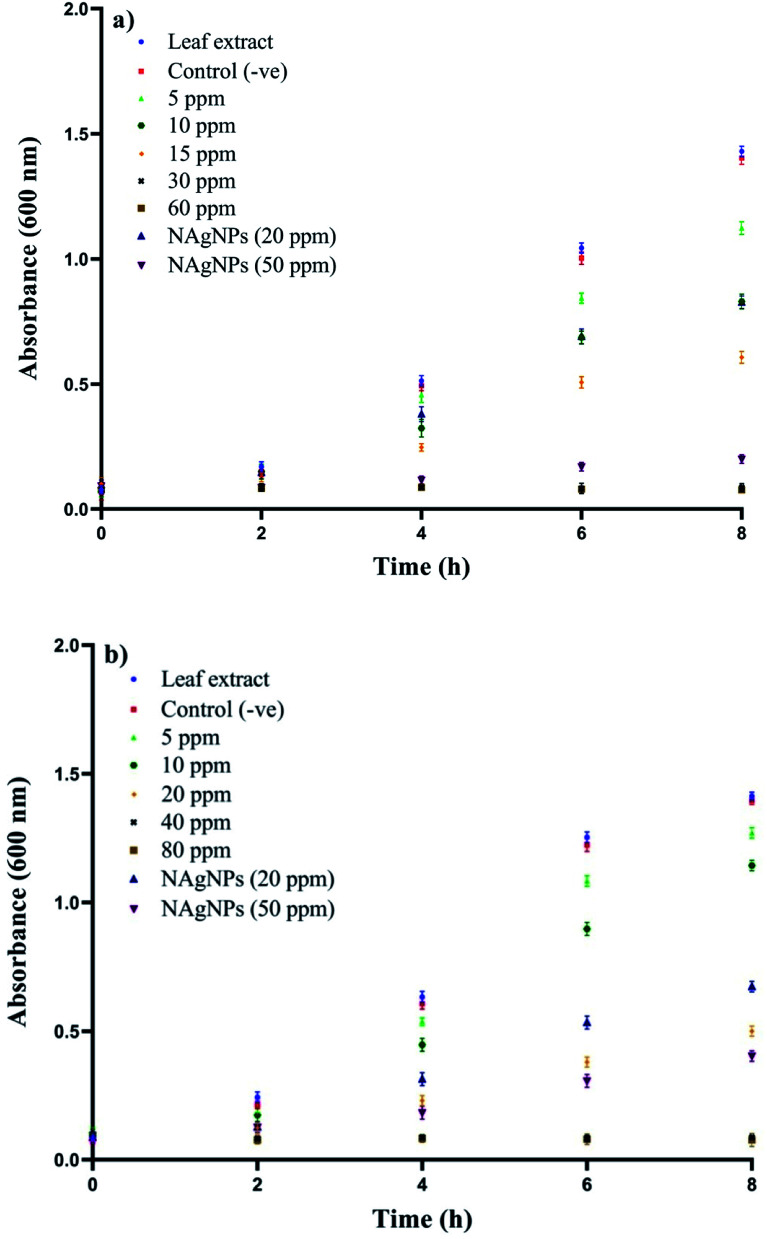
The antibacterial effect of the synthesised AgNPs was tested on bacterial strains ASM and ASE isolated from silkworm. The pure cultures of the test bacteria were sub-cultured in Mueller–Hinton (MH) broths (HiMedia, M391) separately. The overnight cultures of bacterial suspension (1%) were inoculated into fresh MH broth containing a varied concentration of AgNPs (5 ppm, 10 ppm, 15 ppm, 30 ppm and 60 ppm for ASM strain and 5 ppm, 10 ppm, 20 ppm, 40 ppm and 80 ppm for ASE strain, respectively) and kept for incubation at 37 °C with continuous shaking. The growth of bacteria was tracked by calculating the absorbance of the culture media at 600 nm at different time intervals. Minimum inhibitory concentration (MIC), the lowest concentration of AgNPs that inhibits the growth of bacteria, was determined. In addition, the minimum bactericidal concentration (MBC), which is the lowest concentration of antibacterial agent needed to destroy a specific bacterium, was also determined. Both triplicate tests were performed and the mean values recorded. As a negative control, we used MH broth comprising bacterial inoculums without any AgNPs. As a positive control naked AgNPs (with sodium citrate as stabiliser) were used to bring into comparison with bio-molecule functionalized AgNPs. Moreover, the effect of leaf extract on the growth kinetics of the test strain was also checked.
2.9. Cytotoxicity study
2.9.1. Maintenance of cell lines
Cell lines like ACHN (Human, Caucasian, kidney, adenocarcinoma), HEK-293 (human embryonic kidney 293 cells), WRL-68 (Human, liver, embryonic) and HUH-7 (Hepatocellular carcinoma) were procured from NCCS (National Centre for Cell Science) Pune, Maharashtra, India. All adherent cell lines were grown in vitro until they had covered the surface area available or until the medium was depleted of nutrients. DMEM/F-12 (1 : 1 mixture of DMEM and Ham's F-12) medium were used along with glucose, amino acids, and vitamins. The DMEM/F-12 media was augmented with 10% Fetal Bovine Serum (FBS), sodium bicarbonate buffer system to maintain physiological pH in a 5% CO2 environment. The cell lines were routinely sub-cultured in order to avoid the culture dying due to overgrowth. For subculture, the cells were brought into the suspension by proteases, trypsin–EDTA solution or cell scrapers. Cell lines were maintained in original medium at a 1 : 2 split ratio into new Petri dishes. Cell lines were grown aseptically, without stress, and morphology and feasibility of cultures were monitored frequently and cautiously. The cell lines media in the Petri dishes were also examined microscopically for microbial contamination before performing any experiment. This included check-up of unusual pH shifts (red to yellow), turbidity, or particles, fungal colonies through the microscope. Cells numbers were counted by haemocytometer and viability of cells were confirmed by trypan blue exclusion assay before performing MTT assay.
2.9.2. MTT assay
Healthy cells, both for normal (WRL-68 and HEK-293) and cancer cell lines (HUH-7 and ACHN) were seeded in a 96 well microtiter plate with DMEM/F-12 media and kept in an incubator in 5% CO2, at 37 °C for 24 h. On the next day, the sonicated AgNPs sample to aqueous solution of varied concentrations (50, 100, 150, 200 and 250 ppm) was added in each well, except control, and kept for further incubation for 24 h in an incubator containing 5% CO2 at 37 °C in the same condition. The effects of leaf extract and naked AgNPs on the cell lines were also checked. After incubation, media were removed from the well and 10 μL of freshly prepared MTT [3-(4,5 dimethylthiazol-2-yl)-2,5-diphenyl tetrazolium bromide] reagent was added in each well and kept for further incubation for 3 h. The MTT enters the cells and passes into mitochondria during incubation, and it is then reduced into insoluble formazan by dehydrogenase. The insoluble formazan was then solubilised in 50 μL isopropanol and the absorbance was recorded at 620 nm using SPECTROstar Nano BMG LABTECH micro titer plate reader.53 All factors were synchronised before calculating cell viability (e.g. dilution, mixing, sonication time factor, trypsinisation etc.). The percentage of cell viability was calculated as 100 − (Y − X/Y × 100), where Y is the mean optical density of control (untreated cells) and X is the mean optical density of AgNPs treated cells.
2.10. Statistical analysis
The data of DLS, antimicrobial, and cytotoxicity study were analysed using one-way ANOVA and values were expressed as mean ± standard deviation.
3. Results and discussion
3.1. Synthesis and characterisation of bio-molecule functionalized AgNPs
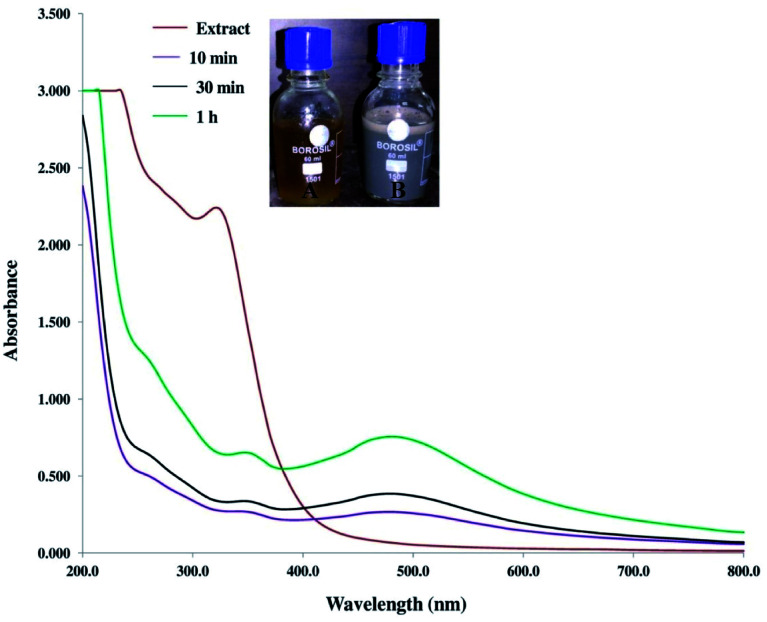
The AgNPs were initially synthesised by the reduction of Ag+ to Ag0, using 25 mL of leaf extract with 50 mL of 10 mM AgNO3 (optimum concentration) and monitored via UV vis-spectroscopy. The formation of AgNPs was confirmed by the solution changing colour from pale yellow to light brown within 10 min. After 1 h the colour was transformed to dark brown which confirmed the completion of the reaction. The spectra of UV-vis showed a surface plasmon resonance (SPR) band at approximately 480 nm (Fig. 1), which is the unique SPR band of AgNPs.54 SPR is attributed to the excitation of electrons at the surface of metallic nanoparticles, displaying a typical pattern of optical absorption in the UV-vis range. For the aqueous leaf extract solution used as the control, the absorption peak was not observed in the same wavelength range. The above observations established the formation of AgNPs. In the plant extract a peak was noted in the range of approximately 290–350 nm, which is a characteristic of natural phenolic acids such as gallic acid. Such active components in plant extract act as reducing and capping agents. When they bind to surface metal, NPs may exhibit a red shift accordingly.16,55
Fig. 1. UV-visible spectra of the mulberry aqueous leaf extract and synthesised AgNPs. Inset (A) and inset (B) show the colour of the leaf extract and biosynthesised AgNPs, respectively.
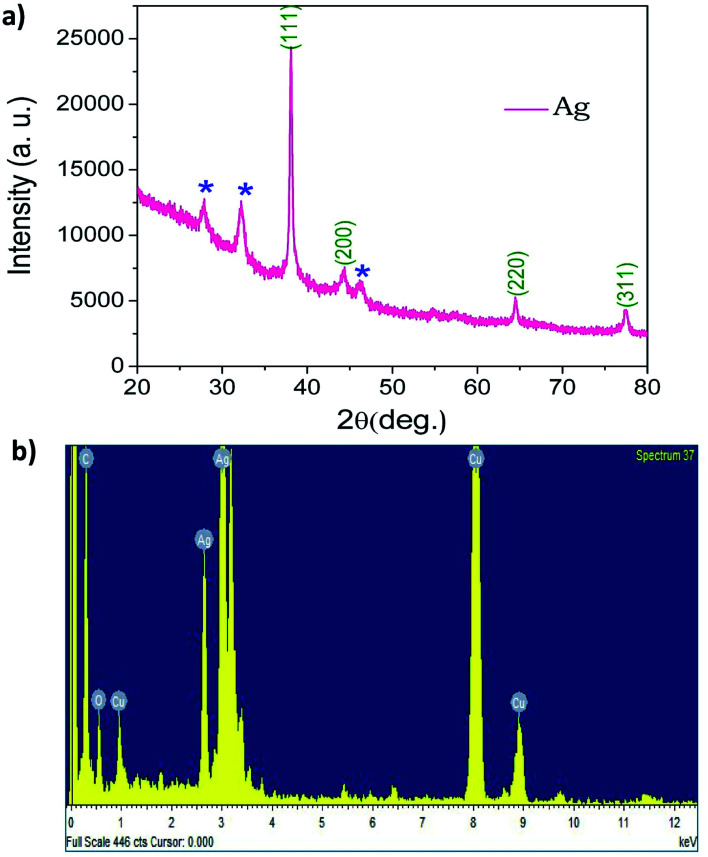
The crystalline structure of AgNPs was determined by XRD analysis. The XRD pattern of biosynthesised AgNPs is presented in Fig. 2a. The presence of peaks on lowered diffraction angles (denoted in asterisks) is due to the existence of phytochemicals in the leaf. The higher intensity peak, indexed as (111) in the XRD pattern, indicates the successful materialisation, and the small size of the crystals.56 The peaks of intensity at the 2θ values of 38.2°, 44.5°, 64.7° and 77.5°, corresponding to the (111), (200), (220), and (311) planes, reveal the face-centred cubic (fcc) nature of AgNPs. To corroborate the chemical analysis, an EDX of the sample was also carried out. The presence of elemental silver in the NPs was proven by a strong absorbance peak at 3 keV (Fig. 2b and Table S1‡). The peak for C and O suggests the bio-functionalization of AgNPs. The strong peak for Cu in EDX is due to the copper grid used in the sample preparation for TEM and EDX.
Fig. 2. (a) XRD spectrum of biosynthesised AgNPs using leaf extract of mulberry (b) EDX spectra of AgNPs showing a strong signal in the silver region.
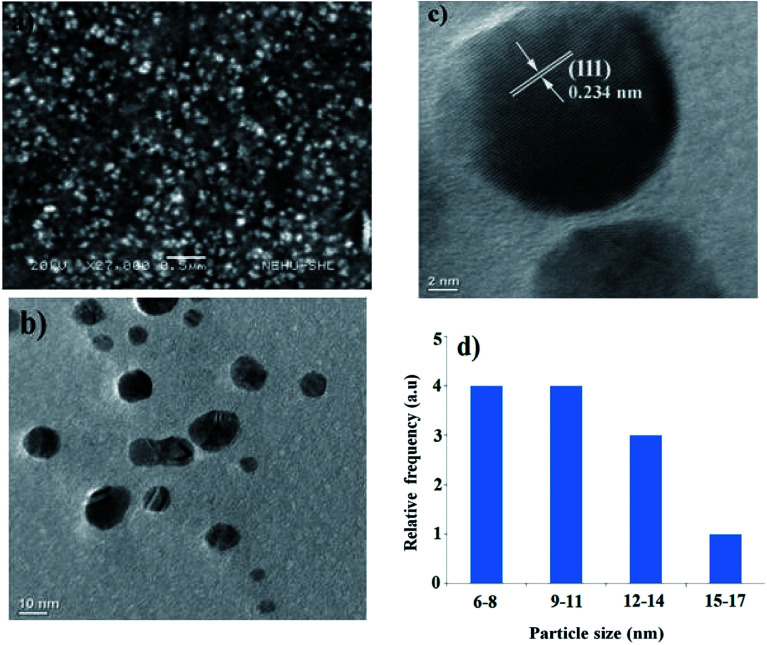
The SEM (Fig. 3a) and TEM (Fig. 3b) micrograph shows that the biosynthesised AgNPs were almost spherical, well dispersed, with an average particle size of 11.8 nm ± 2.8, determined from the histogram of particle size distribution (Fig. 3d). The faded thin coating surrounding the biosynthesised AgNPs confirms its bio-functionalization. The lattice constant with a distance of 0.234 nm revealed in the HR-TEM photograph confirmed the crystalline nature of the AgNPs (Fig. 3c). In our earlier study, we recorded a lattice fringe with a distance of 0.234 nm for green synthesised AgNPs.46 In a separate study, distances of 0.23 nm and 0.235 nm between lattice planes were reported.57,58 AgNPs synthesised using seed extract of Alpinia katsumadai showed a lattice fringe with a distance of 0.238 nm.59 These earlier reports further support our claim.
Fig. 3. (a) SEM image of the synthesised AgNPs (b) TEM image of AgNPs (c) lattice constant of the biosynthesised AgNPs (d) particle size distribution of synthesised AgNPs obtained from TEM micrographs.
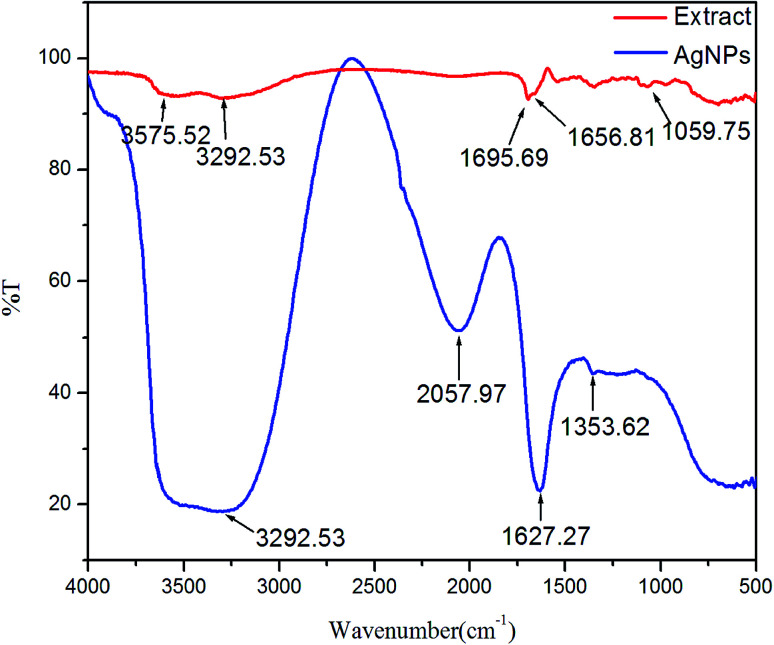
An FT-IR was performed to determine the major functional groups responsible for the reduction of Ag+ to AgNPs. The FT-IR spectrum of leaf extract of mulberry showed sharp absorption bands at 1059, 1656, 1695, 3292 and 3575 cm−1 (Fig. 4). The absorption peaks at 1656 and 1695 cm−1 were assigned to strong symmetric and anti-symmetric stretching vibrations of the carbonyl group of α,β-unsaturated compounds respectively. The broad peaks at 3292 and 3575 cm−1 showed the occurrence of OH stretching in flavonoids, xanthonoids and phenolic compounds, while the band at 1059 cm−1 appeared due to C–O stretching.60,61 The absorption peaks at 1627 cm−1 of AgNPs was due to its carbonyl stretching frequency. The shifting of the stretching frequency of the carboxylic group (COOH) carbonyl from higher (in extract) to lower wavenumber (in AgNPs) is possibly due to the reduction of silver ions (Ag+) by the biomolecules in the leaf. Polar groups like COOH, OH and CO have a high tendency to attach to the surface of the Ag+, causing reduction and stabilisation of Ag+ ions. A similar kind of explanation was also given in an earlier study where synthesis of AgNPs was achieved using Salvia spinosa.16
Fig. 4. FT-IR spectra of leaf extract (red colour) and AgNPs (blue colour).
The DLS study revealed the hydrodynamic size and surface zeta potential of the bio-molecule functionalized AgNPs in a colloidal aqueous environment. Our results show that the AgNPs in MH broth and DMEM/F12 medium had a hydrodynamic size of 25.3 and 50.5 nm, respectively. The average hydrodynamic size and PDI values are summarised in Table S2.‡ Our results show that the hydrodynamic size varied with the composition of the medium. Zeta potential values were found to be −17.3 mV and −25.6 mV at 0 min and −16.8 mV and −24.2 mV after 30 min in MH broth and DMEM/F-12 medium respectively. The high zeta potential indicates its stability with wide-spread electrical surface charge on AgNPs, inhibiting the particles from agglomerating. The PDI values indicated that the systems are fairly monodispersed (Table S2‡). Earlier study revealed a ξ-potential value of −21.7 mV for AgNPs synthesised using orange peel extract,62 while, a zeta potential value of −19.7 mV was recorded in another report on biosynthesis of AgNPs using Trichoderma longibrachiatum.63 Generally, it is believed that values above +30 mV or below −30 mV in a colloid system are electrostatically stable. The size obtained from TEM is generally smaller than DLS, which may be because TEM demands samples in their dry state, whereas, DLS observes the sample in a solution.
3.2. Mass spectrometry
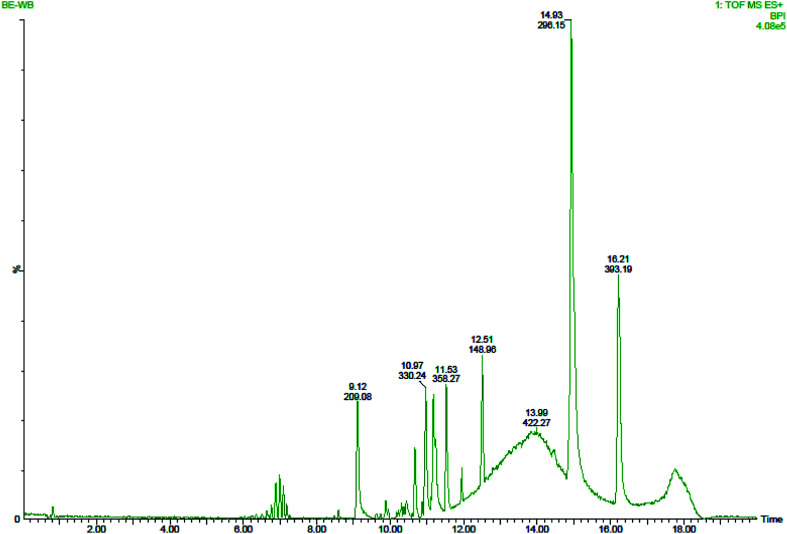
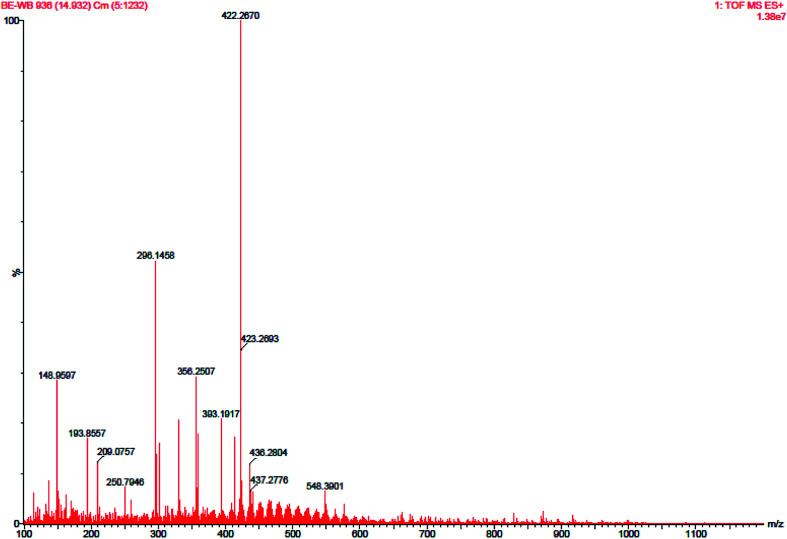
Six peaks were observed from the chromatogram of LC-MS for aqueous leaf extract of mulberry. Some bio-active compounds were identified by comparing the LC retention time (Fig. 5) and ESI-MS spectrometric data (Fig. 6) with those of reference compounds in the database. Owing to the unavailability of authentic standards, some peaks were tentatively assigned by comparing the ESI-MS/MS spectra with those found in previously published articles.64–67 The compounds were assigned and the results are summarised in Table S3.‡ In the aqueous leaf extracts of mulberry, some polyhydroxy compounds were found and matched using examples found in standard literature. Sophora-iso-flavanone A and cyclomorusin fall under the flavonoid family. We successfully identified one polyhydroxy acid; gallic acid, in the leaf extract. Kazinol B is a polyhydroxyflavan (benzopyran derivatives) which was found in the leaf extract. The presence of mangiferin xanthonoid was also revealed by ESI-MS spectra. Apart from these polyhydroxy compounds, a plant steroid, stigmasterol was found in aqueous leaf extracts of mulberry. Earlier study on the phytochemistry revealed the presence of similar bio-active compounds such as gallic acid in the leaf extract of Morus sp.46,64 as well as kazinol B, sophora-iso-flavanone A, cyclomorusin, mangiferin xanthonoid and stigmasterol in the aqueous leaf extract of M. indica L. V1 which further supports our findings.46
Fig. 5. Various components in the aqueous leaf extract of mulberry identified by LC-QTOF/MS.
Fig. 6. HRMS spectra of mulberry leaf extract.
3.3. Possible mechanism for the synthesis of AgNPs
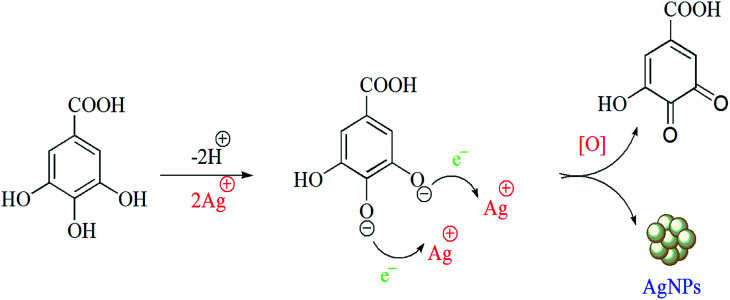
There are only a few reports on the exact mechanism of nanoparticle formation using biological sources. The reduction of Au, Ag, and bimetallic Au core–Ag shell nanoparticles using leaf extract of Azadirachta indica has been reported in the literature.68 In another study, it was reported that several strains of Fusarium oxysporum catalysed the NADPH mediated bio-reduction of AgNO3 to AgNPs.69 The biomolecules bearing naphthoquinones and moieties of anthraquinones have enough redox potential to reduce metal ions and may serve as electron shuttles.70 These reports suggest that biological sources have the potential for nanoparticle synthesis. The LC-MS study of aqueous extract of Morus alba L. S-1635 revealed the presence of some polyhydroxy compounds of the flavonoid and xanthonoid families. Moreover, a few polyhydroxy acid compounds were also found. We hypothesise that the –OH and –COOH groups present in the polyhydroxy compounds, such as gallic acid which revealed by FT-IR and UV-vis spectroscopy, might be responsible for the bio-reduction of silver ions to AgNPs (Scheme 2). Upon the synthesis of AgNPs, gallic acid gets oxidised to its corresponding quinine and absorbed on the surface of NPs via electrostatic interaction between carboxylic acid groups of gallic acid on the surface of the nanoparticles. Additionally, gallic acid molecules on the nanoparticle surface form hydrogen bonds with neighbouring molecules.
Scheme 2. Phytochemical mediated reduction of silver ions and AgNPs synthesis.
3.4. Molecular identification of gut bacteria of infected silkworm
For the screening of bacteria from the gut of infected Bombyx mori L., mannitol salt agar and eosin methylene blue agar were used. Mannitol salt agar is composed of NaCl, mannitol and phenol red as pH indicator. Mannitol salt agar is selective, only allowing the growth of high salt or saline-loving organisms, and differential because of the mannitol fermenter changing the pH indicator colour (red) to yellow (acidic). Non-mannitol fermenters do not change the colour of the medium. Our results showed that bacterium ASM turns the red pH indicator in mannitol salt agar into yellow, which is due to the fermentation of mannitol (Fig. 7a).
Fig. 7. Bacterial colony screened from the gut of infected silkworm on selective media: (a) ASM on mannitol salt agar; (b) ASE on eosin methylene blue agar.
EMB agar plates only allow Gram-negative bacteria to grow on them, because both the eosin and the methylene blue in EMB agar will inhibit the growth of Gram-positive bacteria. The strain ASE produces mucoid with no metallic sheen on EMB agar plates, which confirms that it is Gram-negative (Fig. 7b).
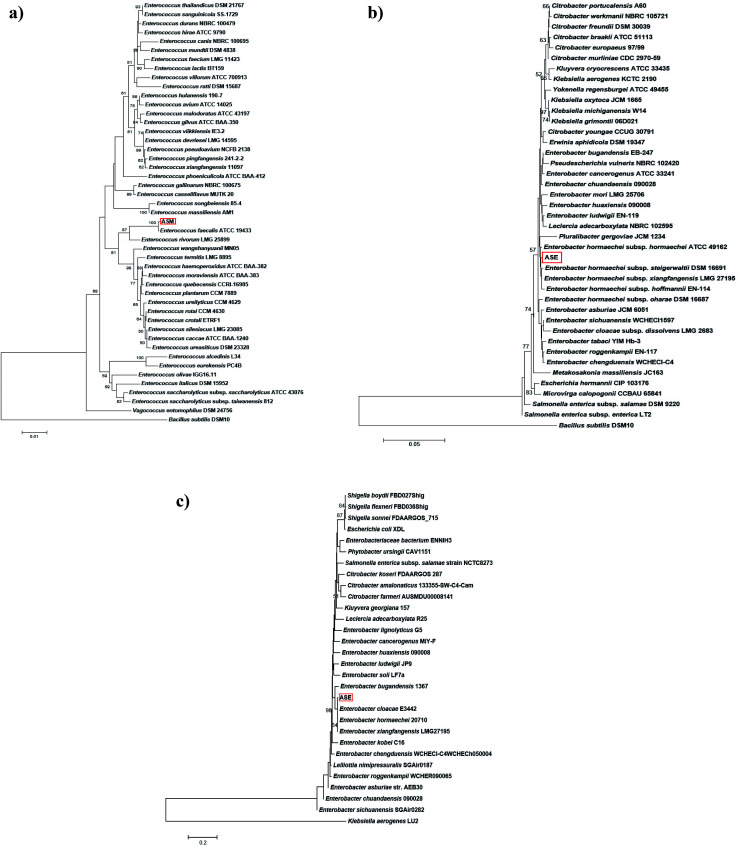
Based on analysis of the phylogenetic tree obtained from the 16S rDNA sequences, strain ASM (NCBI accession no. MT023669) was found to be 100% identical to Enterococcus faecalis ATCC 19433, while strain ASE (NCBI accession no. MT023436) was placed in a separate clade in its phylogenetic tree in spite of having 99.65% similarity with Enterobacter hormaechei subsp. hormaechei ATCC 49162 (Fig. 8a and b). In addition, the phylogenetic tree, obtained from the gyrB gene sequence (nucleotide) reveals isolate ASE to be different from its closest relatives (Fig. 8c).
Fig. 8. (a) Neighbour-joining phylogenetic tree of isolate ASM (NCBI accession no. MT023669) based on 16S rDNA linear sequence. (b) Neighbour-joining phylogenetic tree of isolate ASE (NCBI accession no. MT023436) based on 16S rDNA linear sequence. (c) Neighbour-joining phylogenetic tree of isolate ASE (NCBI accession no. MT023436) based on gyrB gene nucleotide sequence.
3.5. Antibiogram of the test strain
The test strain including Enterobacter hormaechei subsp. hormaechei strain ASE found to be resistant to cell wall inhibitor antibiotics, such as carbenicillin, penicillin, bacitracin, mezlocillin, and cefpodoxime; nucleic acid synthesis inhibitor antibiotics like, novobocin, and nitrofurantoin; and protein synthesis inhibitor antibiotics like oleandomycin and lincomycin, which indicates that isolate ASE is a multi-drug resistant (MDR) bacteria (Table S4‡). Such MDR strains could easily be controlled by the synthesised bio-conjugated AgNPs. Furthermore, as these formulations of AgNPs are made with mulberry leaf extract, they can be easily fed to silkworms, and the desired inhibition of such MDR strains residing in the silkworm gut can be achieved. It was noted that the application of this nano-conjugate can also be widely used against other pathogens or their MDR varieties associated with various diseases.
3.6. Antibacterial activity of AgNPs
The antibacterial activity of the bio-molecule functionalized AgNPs was tested against bacterial strains ASM and ASE, and it was noted that the bacterial growth was inhibited by AgNPs, with dose-dependent effects of the NPs. The minimum inhibitory concentration (MIC) and minimum bactericidal concentration (MBC) values of the AgNPs are shown in Fig. 9. The MBC is the lowest dosage of an antibacterial agent needed to kill 100% of a particular bacterium, which can be measured via broth dilution of MIC tests by sub-culturing to agar plates or broth without any antibacterial agent.71 The growth of Enterococcus faecalis strain ASM in MH broth was completely inhibited within 2 h when grown with 30 ppm of AgNPs, and no growth was noticed at 60 ppm, even after overnight incubation. Therefore, the concentration of 30 ppm was considered the MIC for the ASM strain and the concentration of 60 ppm was considered the MBC (Fig. 9a). Similarly, the growth of Enterobacter hormaechei subsp. hormaechei strain ASE in MH broth was inhibited within 2 h in the presence of 40 ppm of AgNPs and no growth was observed at 80 ppm, even after overnight incubation. Therefore, the concentration of 40 ppm was considered the MIC for the ASE strain and the concentration of 80 ppm was considered the MBC (Fig. 9b).
Fig. 9. Growth kinetics of the isolates in MH broth with varied concentrations of the AgNPs: (a) Enterococcus faecalis strain ASM (b) Enterobacter hormaechei subsp. hormaechei strain ASE. The results shown are the mean ± SD of three separate experiments done in triplicate.
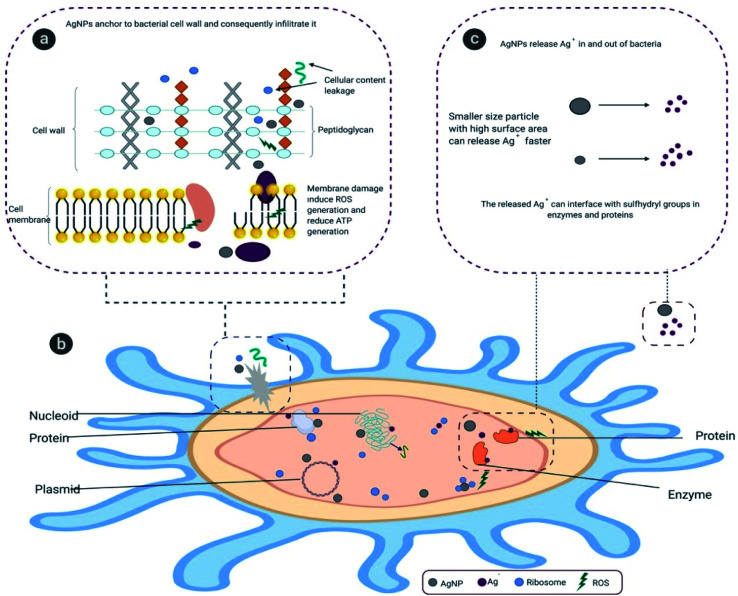
In an earlier study the MIC value of chemically synthesised nanosilver with an average particle size of 35 nm was recorded at 50 ppm against Enterococcus faecalis.72 In another study, biogenic AgNPs of the same size showed a MIC value of 100 ppm against Enterobacter aerogenes MTCC 2824.73 Our comparative antibacterial study with naked AgNPs showed that bio-molecule functionalized AgNPs are more potent in hindering bacterial growth compared to naked AgNPs (Fig. 9). It was also revealed that the aqueous leaf extract did not restrain the growth of the tested bacteria. In general, the toxicity of AgNPs is determined by the dissolution, or ionisation from Ag° to Ag+. Therefore, microbial cell death and the disruption of normal cell function are enhanced by internalisation of Ag+ and cellular bio-molecules disruption. In oxygenic condition the AgNPs dissolute and release Ag+, contributing towards the formation of intracellular reactive oxygen species (ROS), which damage protein, DNA, RNA, lipid, cofactor and ultimately lead to cell death (Scheme 3).74,75
Scheme 3. Mechanism of AgNPs dissolution and formation of ROS.
3.7. Cytotoxicity study
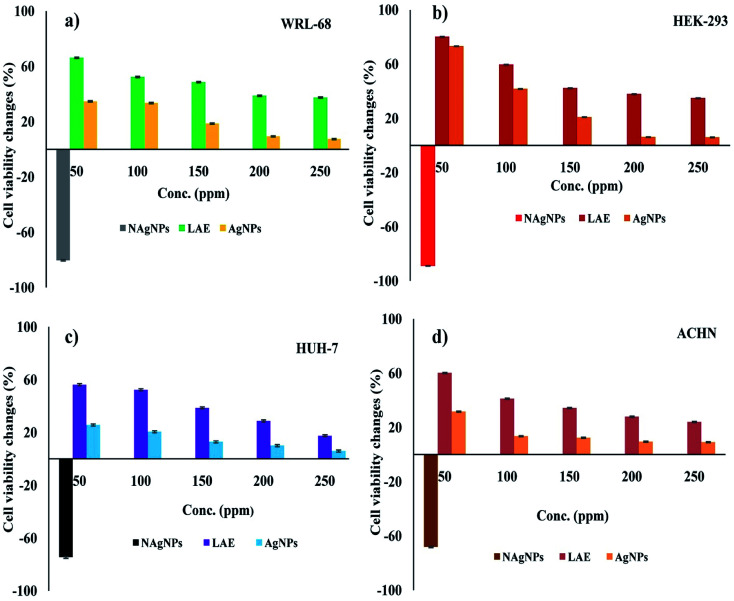
To evaluate the effect of AgNPs on humans, human kidney and liver cell lines were screened to study the effect of nanoparticles on growth and cell population. To determine the viability of cells in the presence of AgNPs, a MTT assay was employed on WRL-68, HEK-293, HUH-7, and ACHN cell lines (Fig. 10). The MTT assay relies on the mitochondrial respiration system and secondarily assists in evaluating the cellular energy capacity of a cell. The MTT assay was applied to measure the viability of AgNP-treated cells. MTT is a yellow dye, which is reduced by a cellular enzyme, oxido-reductase, where nicotinamide adenine dinucleotide phosphate (NADPH) acts as electron donor and produces a blue product, formazan. This transformation is only feasible in viable cells; thus, the amount of blue formazan is proportional to the number of these viable cells. We found interesting results in the effect of the presence of biosynthesised AgNPs on human cell lines: our findings display no remarkable toxicity nor any notable difference in cell viability in the presence of both aqueous leaf extract as well as bio-molecule functionalized AgNPs with respect to the untreated control. The toxicity of bio-molecule functionalized AgNPs was lower than the naked AgNPs (at 50 ppm) indicating its biocompatibility (Fig. 10). Our study revealed that the bio-molecule functionalized AgNPs specifically damaged prokaryotic cells which may be due to differences in membrane structure between prokaryotic and eukaryotic cells. The endocytosis of AgNPs leads to its dissolution with release of Ag+ in the cytosol, which causes impairment of metabolic and cell cycle mechanisms.74,75 In an earlier report, bio-conjugated AgNPs showed cytotoxicity against a cancerous cell line (MCF-7) without affecting the normal cell line (L-929),whereas, chemically synthesised AgNPs displayed a cytotoxic effect on both.76
Fig. 10. Cell viability analyses using MTT assay in the presence of leaf extract, AgNPs and naked AgNPs (NAgNPs) on human cell lines WRL-68, HEK-293, ACHN, and HUH-7. The results shown are the mean ± SD of three separate experiments done in triplicate.
4. Conclusions
In brief, the present study established an eco-friendly, rapid, one-pot green approach to the synthesis of bio-molecule functionalized AgNPs using aqueous leaf extract of Morus alba L. S-1635. Gallic acid (a polyhydroxy acid) in the leaf extract might be responsible for the reduction of Ag+ into AgNPs. The bio-molecule functionalized AgNPs exhibited promising antibacterial activity against MDR bacteria isolated from the gut of natural infected silkworm. There is no significant difference in cell viability of WRL-68, HEK-293, ACHN, and HUH-7 in the presence of bio-functionalized AgNPs, suggesting its potential candidate for biomedical applications. Thus, our study opened up an unparallel prospect for new bio-fabricated advanced nanomaterial for combating silkworm pathogens to increase the quality of industrial culture conditions of domesticated silkworm production to optimise the silk fibre productivity.
Author's contribution
SS, BS, KB, TKJ, DB, PD, RM, AS, SS and AK designed and performed the experiments. AKD helps in the interpretation of XRD. OLF analysed the results of LC-MS. AK performed cytotoxicity assay. AM, SM, IO, AKM and IAI guided in all experiments and wrote the manuscript with the help of other authors.
Conflicts of interest
There are no conflicts to declare.
Supplementary Material
Acknowledgments
The authors are grateful to Advanced Instrumentation Research Facility-JNU, India, Sophisticated Analytical Instrument Facility-NEHU, India and Central Instrumentation Facility-BIT Mesra, India for providing instrumentation facility.
Dedicated to Prof. Syed Sirajul Islam, Retired Professor of Department of Chemistry and Chemical Technology, Vidyasagar University for his substantial contribution in the field of Carbohydrate Chemistry.
Electronic supplementary information (ESI) available. See DOI: 10.1039/d0ra03451g
Notes and references
- Behravan M. Panahi A. H. Naghizadeh A. Ziaee M. Mahdavi R. Mirzapour A. Int. J. Biol. Macromol. 2019;124:148–154. doi: 10.1016/j.ijbiomac.2018.11.101. [DOI] [PubMed] [Google Scholar]
- Roy A. Bulut O. Some S. Mandal A. K. Yilmaz M. D. RSC Adv. 2019;9:2673–2702. doi: 10.1039/C8RA08982E. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cushing B. L. Kolesnichenko V. L. O C. J. Connor Chem. Rev. 2004;104:3893–3946. doi: 10.1021/cr030027b. [DOI] [PubMed] [Google Scholar]
- Abou El-Nour K. M. M. Eftaiha A. Al-Warthan A. Ammar R. A. A. Arabian J. Chem. 2010;3:135–140. doi: 10.1016/j.arabjc.2010.04.008. [DOI] [Google Scholar]
- Aritonang H. F. Koleangan H. Wuntu A. D. Int. J. Microbiol. 2019;2019 doi: 10.1155/2019/8642303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan M. Z. H. Tareq F. K. Hossen M. A. Roki M. N. A. M. J. Eng. Sci. Technol. 2018;13:158–166. [Google Scholar]
- Budak E. Aykut S. Paşaoğlu M. E. Ünlü C. Mater. Today Commun. 2020;24:100975. doi: 10.1016/j.mtcomm.2020.100975. [DOI] [Google Scholar]
- Rogers K. R. Navratilova J. Stefaniak A. Bowers L. Knepp A. K. Al-Abed S. R. Potter P. Gitipour A. Radwan I. Nelson C. Bradham K. D. Sci. Total Environ. 2018;619–620:1375–1384. doi: 10.1016/j.scitotenv.2017.11.195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calderón-Jiménez B. Johnson M. E. Montoro Bustos A. R. Murphy K. E. Winchester M. R. Vega Baudrit J. R. Front. Chem. 2017;5:6. doi: 10.3389/fchem.2017.00006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y. Tang R. C. J. Photochem. Photobiol., B. 2018;178:463–471. doi: 10.1016/j.jphotobiol.2017.12.003. [DOI] [PubMed] [Google Scholar]
- Firdhouse M. J. Lalitha P. J. Nanotechnol. 2015;2015 doi: 10.1155/2015/829526. [DOI] [Google Scholar]
- Rus A. Leordean V.-D. Berce P. MATEC Web Conf. 2017;137:07007. doi: 10.1051/matecconf/201713707007. [DOI] [Google Scholar]
- Iravani S. Korbekandi H. Mirmohammadi S. V. Zolfaghari B. Results Pharma Sci. 2014;9:385–406. [PMC free article] [PubMed] [Google Scholar]
- Lee S. H. Jun B. H. Int. J. Mol. Sci. 2019;20:865. doi: 10.3390/ijms20040865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moodley J. S. Krishna S. B. N. Pillay K. Sershen Govender P. Adv. Nat. Sci.: Nanosci. Nanotechnol. 2018;9:015011. [Google Scholar]
- Pirtarighat S. Ghannadnia M. Baghshahi S. J. Nanostruct. Chem. 2019;9:1–9. doi: 10.1007/s40097-018-0291-4. [DOI] [Google Scholar]
- Iravani S. Int. Scholarly Res. Not. 2014;2014:1–14. doi: 10.1155/2014/359316. doi: 10.1155/2014/359316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X.-F. Liu Z.-G. Shen W. Gurunathan S. Int. J. Mol. Sci. 2016;17:1534. doi: 10.3390/ijms17091534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krithiga N. Rajalakshmi A. Jayachitra A. Journal of Nanoscience. 2015;2015 doi: 10.1155/2015/928204. doi: 10.1155/2015/928204. [DOI] [Google Scholar]
- Jha A. K. Prasad K. Prasad K. Kulkarni A. R. Colloids Surf., B. 2009;73:219–223. doi: 10.1016/j.colsurfb.2009.05.018. [DOI] [PubMed] [Google Scholar]
- Rautela A. Rani J. Das M. D. J. Anal. Sci. Technol. 2019;10:1–10. doi: 10.1186/s40543-018-0160-2. [DOI] [Google Scholar]
- Some S. Sen I. K. Mandal A. Aslan T. Ustun Y. Yilmaz E. Ş. Katı A. Demirbas A. Mandal A. K. Ocsoy I. Mater. Res. Express. 2019;6:12001. doi: 10.1088/2053-1591/aae23e. [DOI] [Google Scholar]
- Chinnasamy G. Chandrasekharan S. Bhatnagar S. Int. J. Nanomed. 2019;14:9823–9836. doi: 10.2147/IJN.S231340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phull A.-R. Ali A. Abbasi S. Zia M. Khaskheli M. H. Haq I. U. Kamal M. A. Med. Chem. 2020;16:495–506. doi: 10.2174/1573406415666191111143213. [DOI] [PubMed] [Google Scholar]
- Baruah D. Yadav R. N. S. Yadav A. Das A. M. J. Photochem. Photobiol., B. 2019;201:111649. doi: 10.1016/j.jphotobiol.2019.111649. [DOI] [PubMed] [Google Scholar]
- Qayyum S. Oves M. Khan A. U. PLoS One. 2017;12:e0181363. doi: 10.1371/journal.pone.0181363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Datta R. K., FAO Electronic conference on mulberry for animal production, 2000, http://www.fao.org/AG/AGa/AGAP/FRG/Mulberry/Papers/HTML/Datta.htm, accessed February 2020 [Google Scholar]
- Chowdhury P. Setua G. Kar N. Ghosh A. Uttar Pradesh J. Zool. 2011;31:261–274. [Google Scholar]
- Andallu B. Suryakantham V. Srikanthi B. L. Reddy G. K. Clin. Chim. Acta. 2001;314:47–53. doi: 10.1016/S0009-8981(01)00632-5. [DOI] [PubMed] [Google Scholar]
- Andallu B. Varadacharyulu N. C. Clin. Chim. Acta. 2003;338:3–10. doi: 10.1016/S0009-8981(03)00322-X. [DOI] [PubMed] [Google Scholar]
- Singh J. Singh N. Rathi A. Kukkar D. Rawat M. J. Nanostruct. 2017;7:134–140. [Google Scholar]
- Adavallan K. Krishnakumar N. Adv. Nat. Sci.: Nanosci. Nanotechnol. 2014;5:025018. [Google Scholar]
- Chen B. Du K. Sun C. Vimalanathan A. Liang X. Li Y. Wang B. Lu X. Li L. Shao Y. ISME J. 2018;12:2252–2262. doi: 10.1038/s41396-018-0174-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Omar S. Fathy D. Journal of Agricultural Chemistry and Biotechnology. 2016;7:213–218. doi: 10.21608/jacb.2016.40892. [DOI] [Google Scholar]
- Dong H.-L. Zhang S.-X. Tao H. Chen Z.-H. Li X. Qiu J.-F. Cui W.-Z. Sima Y.-H. Cui W.-Z. Xu S.-Q. Sci. Rep. 2017;7:10972. doi: 10.1038/s41598-017-11592-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tekin O. K. Sezer D. Soganci K. Tunca R. I. Turkish Journal of Agricultural and Natural Sciences. 2014;14:450–453. [Google Scholar]
- Bhat I. A. Buhroo Z. I. Bhat M. A. Int. J. Entomol. Res. 2017;2:1–9. [Google Scholar]
- Berche P. Clin. Microbiol. Infect. 2012;18:1–6. doi: 10.1111/j.1469-0691.2012.03945.x. [DOI] [PubMed] [Google Scholar]
- Tao H.-P. Shen Z.-Y. Zhu F. Xu X.-F. Tang X.-D. Xu L. Curr. Microbiol. 2011;62:876–883. doi: 10.1007/s00284-010-9796-x. [DOI] [PubMed] [Google Scholar]
- Ganeshprabu P. Selvi S. Mathivanan V. Int. J. Pure Appl. Math. 2013;3:89–93. [Google Scholar]
- Nesa J. Sadat A. Buccini D. F. Kati A. Mandal A. K. Franco O. L. RSC Adv. 2020;10:512–523. doi: 10.1039/C9RA06864C. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Y. L., FAO Agricultural Services Bulletin, FAO, Rome, 1991, pp. 27–35 [Google Scholar]
- Cappellozza S. Saviane A. Tettamanti G. Squadrin M. Vendramin E. Paolucci P. Franzetti E. Squartini A. J. Invertebr. Pathol. 2011;106:386–393. doi: 10.1016/j.jip.2010.12.007. [DOI] [PubMed] [Google Scholar]
- Shashidhar K. R. Thulasiram K. Noorulla H. J. Pharmacogn. Phytochem. 2018;7:2720–2724. [Google Scholar]
- Manna D. K. Mandal A. K. Sen I. K. Maji P. K. Chakraborti S. Chakraborty R. Islam S. S. Int. J. Biol. Macromol. 2015;80:455–459. doi: 10.1016/j.ijbiomac.2015.07.028. [DOI] [PubMed] [Google Scholar]
- Some S. Bulut O. Biswas K. Kumar A. Roy A. Sen I. K. Mandal A. Franco O. L. İnce İ. A. Neog K. Das S. Pradhan S. Dutta S. Bhattacharjya D. Saha S. Das Mohapatra P. K. Bhuimali A. Unni B. G. Kati A. Mandal A. K. Yilmaz M. D. Ocsoy I. Sci. Rep. 2019;9:14839. doi: 10.1038/s41598-019-50906-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maji A. Beg M. Mandal A. K. Das S. Jha P. K. Hossain M. Process Biochem. 2017;60:59–66. doi: 10.1016/j.procbio.2017.05.022. [DOI] [Google Scholar]
- Marmur J. A. J. Mol. Biol. 1961;3:208–218. doi: 10.1016/S0022-2836(61)80047-8. [DOI] [Google Scholar]
- Turner S. Pryer K. M. Miao V. P. W. Palmer J. D. J. Eukaryotic Microbiol. 1999;46:327–338. doi: 10.1111/j.1550-7408.1999.tb04612.x. [DOI] [PubMed] [Google Scholar]
- Yamamoto S. Harayama S. Appl. Environ. Microbiol. 1995;61:1104–1109. doi: 10.1128/AEM.61.3.1104-1109.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura M. J. Mol. Evol. 1980;16:111–120. doi: 10.1007/BF01731581. [DOI] [PubMed] [Google Scholar]
- Bauer A. W. Kirby W. M. M. Sherris J. C. Turck M. Am. J. Clin. Pathol. 1966;36:493–496. doi: 10.1093/ajcp/45.4_ts.493. [DOI] [PubMed] [Google Scholar]
- Denizot F. Lang R. J. Immunol. 1986;89:271–277. doi: 10.1016/0022-1759(86)90368-6. [DOI] [PubMed] [Google Scholar]
- Jiang Z. Wen G. Luo Y. Zhang X. Liu Q. Liang A. Sci. Rep. 2014;4:5323. doi: 10.1038/srep05323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masek A. Latos-Brozio M. Chrzescijanska E. Podsedek A. Forests. 2019;10:988. doi: 10.3390/f10110988. [DOI] [Google Scholar]
- Bhagat M. Anand R. Datt R. Gupta V. Arya S. J. Inorg. Organomet. Polym. 2019;29:1039–1047. doi: 10.1007/s10904-018-0994-5. [DOI] [Google Scholar]
- Khan M. J. Kumari S. Shameli K. Selamat J. Sazili A. Q. Materials. 2019;12:2382. doi: 10.3390/ma12152382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Philip D. Unni C. Aromal S. A. Vidhu V. K. Spectrochim. Acta, Part A. 2011;78:899–904. doi: 10.1016/j.saa.2010.12.060. [DOI] [PubMed] [Google Scholar]
- He Y. Wei F. Ma Z. Zhang H. Yang Q. Yao B. Huang Z. Li J. Zeng C. Zhang Q. RSC Adv. 2017;7:39842–39851. doi: 10.1039/C7RA05286C. [DOI] [Google Scholar]
- Kong J. Yu S. Acta Biochim. Biophys. Sin. 2007;39:549–559. doi: 10.1111/j.1745-7270.2007.00320.x. [DOI] [PubMed] [Google Scholar]
- Prathnaa T. C. Chandrasekaran N. Raichur A. M. Mukherjee A. Colloids Surf., B. 2011;82:152–159. doi: 10.1016/j.colsurfb.2010.08.036. [DOI] [PubMed] [Google Scholar]
- Skiba M. I. Vorobyova V. I. Adv. Mater. Sci. Eng. 2019;2019:8306015. [Google Scholar]
- Elamawi R. M. Al-Harbi R. E. Hendi A. A. Egypt J. Biol. Pest Co. 2018;28:28. doi: 10.1186/s41938-018-0028-1. [DOI] [Google Scholar]
- Flaczyk E. Kobus-Cisowska J. Przeor M. Korczak J. Remiszewski M. Korbas E. Buchowski M. Agric. Sci. 2013;4:141–147. [Google Scholar]
- Sohn H. Y. Son K. H. Kwon C. S. Kwon G. S. Kang S. S. Phytomedicine. 2004;11:666–672. doi: 10.1016/j.phymed.2003.09.005. [DOI] [PubMed] [Google Scholar]
- Imran M. Arshad M. S. Butt M. S. Kwon J. H. Arshad M. U. Sultan M. T. Lipids Health Dis. 2017;16:84. doi: 10.1186/s12944-017-0449-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zambakhidze N. E. Sulaberidze K. V. Mzhavanadze V. V. Tsiklauri G. C. Appl. Biochem. Microbiol. 2005;41:404–406. doi: 10.1007/s10438-005-0069-6. [DOI] [Google Scholar]
- Shankar S. S. Rai A. Ahmad A. Sastry M. J. Colloid Interface Sci. 2004;275:496–502. doi: 10.1016/j.jcis.2004.03.003. [DOI] [PubMed] [Google Scholar]
- Durán N. Marcato P. D. Alves O. L. De Souza G. I. H. Esposito E. J. Nanobiotechnol. 2005;3:8. doi: 10.1186/1477-3155-3-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newman D. K. Kolter R. Nature. 2000;405:94–97. doi: 10.1038/35011098. [DOI] [PubMed] [Google Scholar]
- Qais F. A. Shafiq A. Khan H. M. Husain F. M. Khan R. A. Alenazi B. Alsalme A. Ahmad I. Bioinorg. Chem. Appl. 2019;2019 doi: 10.1155/2019/4649506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lotfi M. Vosoughhosseini S. Ranjkesh B. Khani S. Saghiri M. Zand V. Afr. J. Biotechnol. 2011;10:6799–6803. [Google Scholar]
- Ojha S. Sett A. Bora U. Adv. Nat. Sci.: Nanosci. Nanotechnol. 2017;8:035009. [Google Scholar]
- Maurer-Jones M. A. Mousavi M. P. S. Chen L. D. Bühlmann P. Haynes C. L. Chem. Sci. 2013;4:2564–2572. doi: 10.1039/C3SC50320H. [DOI] [Google Scholar]
- Molleman B. Hiemstra T. Langmuir. 2015;31:13361–13372. doi: 10.1021/acs.langmuir.5b03686. [DOI] [PubMed] [Google Scholar]
- Khorrami S. Zarrabi A. Khaleghi M. Danaei M. Mozafari M. R. Int. J. Nanomed. 2018;13:8013–8024. doi: 10.2147/IJN.S189295. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.