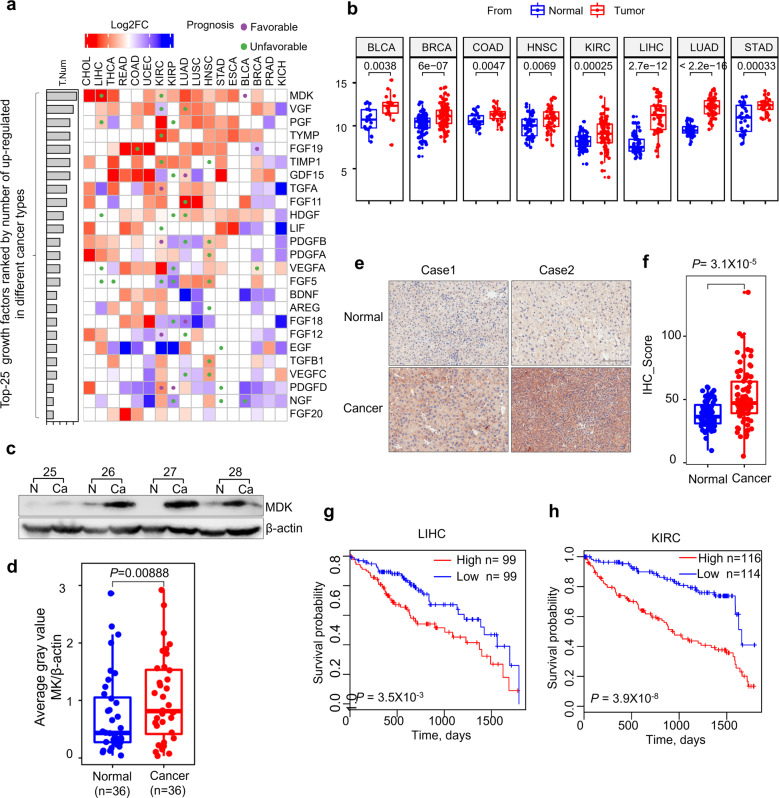
Fig. 5. Midkine expression is upregulated in cancer.
a Pan-cancer evaluation of the expression and prognostic impact of growth factors. The color of each rectangle represents the log2 transformed fold change (Log2FC) of the mRNA expression for the corresponding growth factor between tumor and normal tissues, and the white rectangles indicate either a Log2FC value equal 0 or differences between tumor and normal tissues that are not significant (linear model approach of limma, P > 0.01). The purple and green circles represent high gene expression correlated with good and poor prognosis, respectively (log rank test, P < 0.01). The left bars represent the numbers of cancer types in which a growth factor is upregulated in the tumor tissues compared with the normal tissues (P < 0.01, log2FC > 1). The growth factors are ranked by the numbers, and only the top 25 factors are shown. b Boxplots of the differences in MDK expression in paired normal and tumor tissues of eight types of cancers. The centers of the boxes represent the median values. The bottom and top boundaries of the boxes represent the 25th and 75th percentiles, respectively. The whiskers indicate 1.5-fold of the interquartile range. The dots represent points falling outside this range. The paired P-values were calculated based on Wilcox tests. c–d The expression of MDK in 36 pairs of matched adjacent nontumor (NT) and cancer (Ca) tissues as detected by Western blotting (c), and the distribution of MDK expression in both the NT and Ca samples as represented by boxplots with the expression value normalized by ImageJ software (d). e–f Immunohistochemical staining of MDK in representative adjacent nontumor and HCC specimens (e) and boxplots of the distributions of MDK expression status in 75 paired paraffin-embedded tissues (f). Scale bar, 200 μm. g–h Kaplan–Meier survival curves of LIHC (g) and KIRC (h) patients with data stratified by the expression levels obtained from the TCGA database.