| 1 |
Huperzine A |
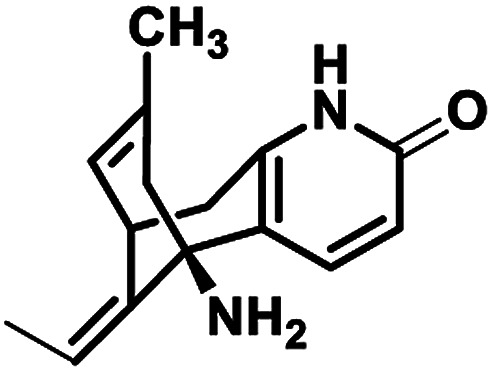
|
−6.970 |
Met631(A), Val634(A), Ala635(A), Phe597(B), Leu626(B), Ala627(B), Tyr629(B), Val634(C), Ala635(C), Ala622(D), Val623(D), Leu626(D), Ala627(D) |
| 2 |
NM14 |

|
−7.475 |
Met631(A), Val634(A), Ala635(A), Phe597(B), Val623(B), Leu626(B), Ala627(B), Tyr629(B), Val634(C), Val623(D), Leu626(D), Ala627(D) |
| 3 |
NM16 |

|
−7.542 |
Met631(A), Val634(A), Ala635(A), Leu626(B), Ala627(B), Tyr629(B), Val634(C), Ala635(C), Val623(D), Leu626(D), Ala627(D), Ala631(D) |
| 4 |
NM20 |
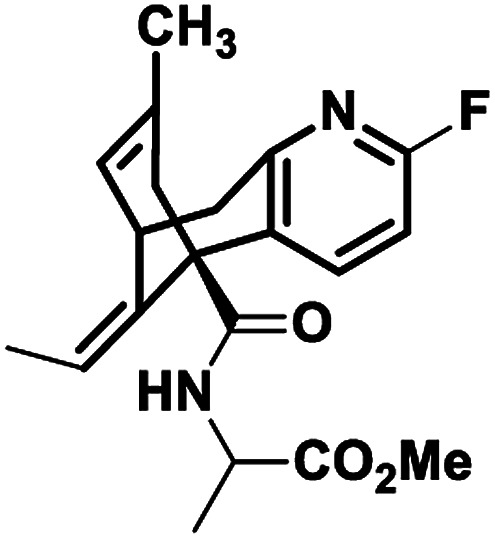
|
−7.722 |
Met631(A), Val634(A), Ala635(A), Val623(B), Leu626(B), Ala627(B), Tyr629(B), Val634(C), Ala622(D), Val623(D), Leu626(D), Ala627(D) |
| 5 |
NM22 |

|
−7.558 |
Met631(A), Val634(A), Ala635(A), Val623(B), Leu626(B), Ala627(B), Val634(C), Ala635(C), Val623(D), Leu626(D), Ala627(D) |
| 6 |
NM28 |
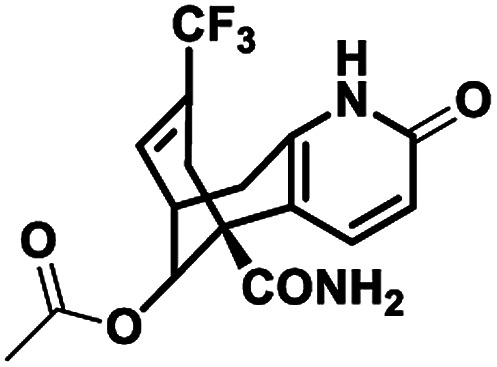
|
−7.830 |
Val634(A), Phe597(B), Val623(B), Leu626(B), Val634(C), Val623(D), Leu626(D), Ala627(D) |
| 7 |
MK-801 |

|
−7.365 |
Met631(A), Val634(A), Ala635(A), Leu626(B), Ala627(B), Tyr629(B), Val623(D), Leu626(D), Ala627(D) |







