Figure 3.
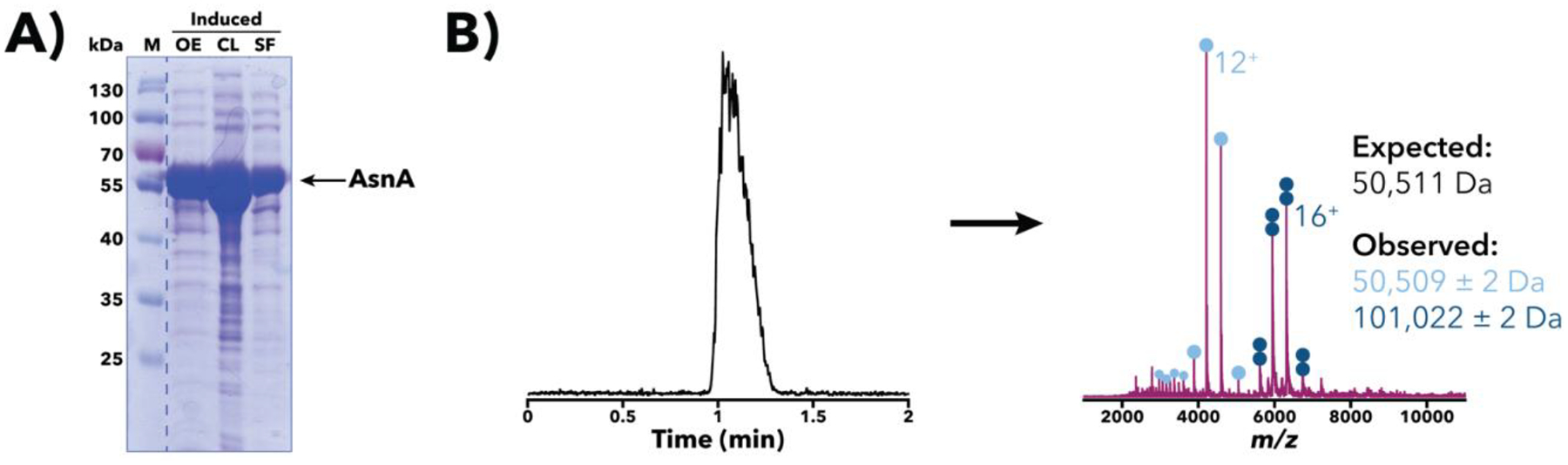
Characterization of a Salmonella AsnA overexpression sample. (A) SDS-PAGE [10% (w/v) polyacrylamide] analysis of a post-induction sample at sequential stages of processing. Despite an expected mass of ~50.5 kDa, the apparent AsnA band runs slower than the 55 kDa marker, an example of aberrant protein migration in SDS-PAGE. Gel image has been spliced at the dotted line to only show relevant samples. M, size markers; OE, whole-cell sample following overexpression; CL, whole-cell crude lysate; SF, soluble fraction post-centrifugation. (B) IMAC-OBE-nMS analysis of the soluble fraction using a 500 mM ammonium acetate mobile phase. A representative total ion chromatogram (left) and mass spectrum (right; IST 100 V, HCD CE 5 eV) are shown. The expected mass noted above accounts for loss of the N-terminal methionine, a common post-translational modification (Ben-Bassat et al., 1987; Giglione et al., 2004). Charge state distributions for monomeric and dimeric AsnA species are indicated with light and dark blue circles, respectively, and the main charge state for each species is labeled. The bimodal charge state distribution for the monomer, with its low intensity, higher charge state peaks marked with smaller light blue circles, suggests that there is partially unfolded monomer present in the sample. The y-axis (not shown) represents relative intensity.
