Figure 1.
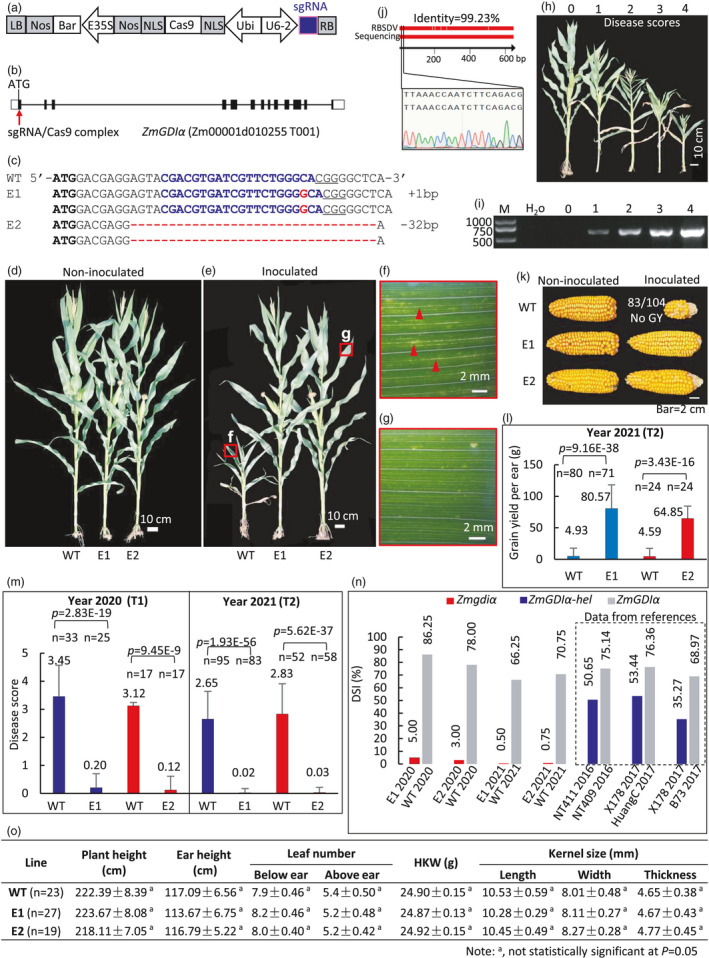
Genome editing ZmGDIα confers resistance against MRDD without agronomic penalty. (a) Constructed vector. (b) The designed target site. (c) Genotype of the selected homozygous mutations. (d, e) Comparison of non‐inoculated (d) and inoculated (e) plants. The leaves in the red rectangles in (e) are magnified to show MRDD symptoms in the WT (f) and E2 (g). (f, g) Typical waxy enation symptom of MRDD is seen on the WT (f) but not in mutant (g). (h) Representative plants with scored MRDD severity from 0 to 4 from resistance to susceptibility. (i) RT‐PCR detection of RBSDV showing that the virus titre is proportional to scored MRDD severity. (j) Verification of RBSDV identity by sequencing RT‐PCR amplicon in (i). (k, l) Comparison of representative maize ears (k) and grain yield (l) in the field. Eighty‐three out of 104 inoculated WT plants produced no grain. (m) Disease scores across years 2020 and 2021. (n) Comparison of DSI observed in the null mutants and the previously reported ZmGDI‐hel. NT411/NT409 and X178/Huang C data were reproduced from Liu C. et al. (2020). X178/B73 data were reproduced from Liu Q. et al. (2016). DSI (%) = ∑(disease score × number of plants with this disease score) × 100 / (maximum disease score × total number of plants). (o) The edited lines show no agronomic penalty without MRDD infection in the field. The differences were not statistically significant at P = 0.05.
