Figure 1.
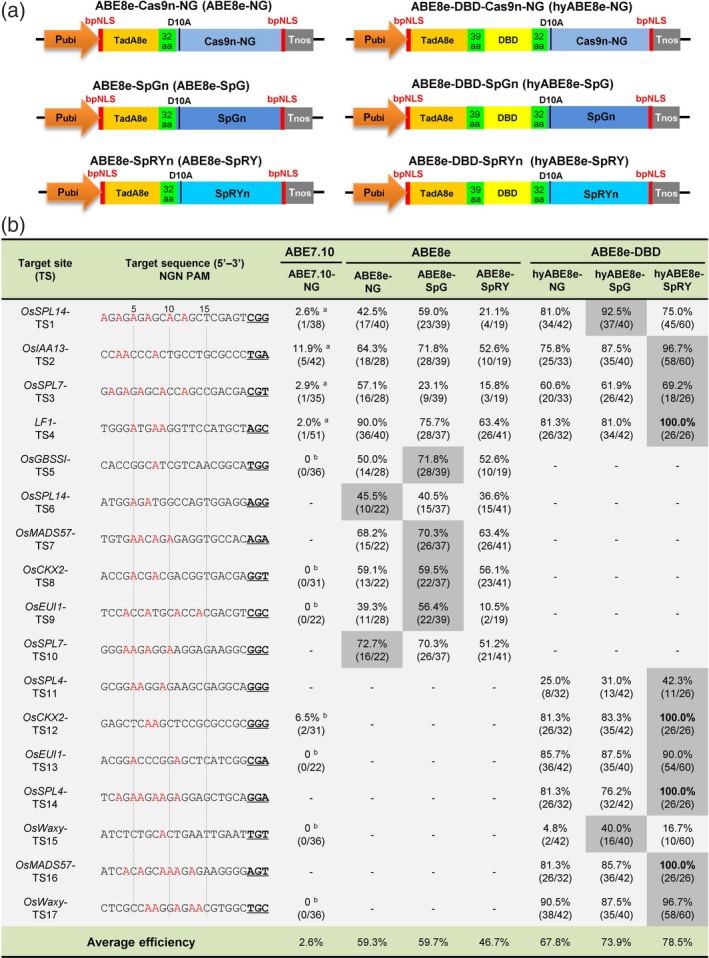
PhieABEs allow efficient A‐to‐G conversion at NGN‐PAM target sites in rice. (a) Schematic diagrams of basic ABE8es (ABE8e‐NG, ABE8e‐SpG and ABE8e‐SpRY) and PhieABE (hyABE8e‐NG, hyABE8e‐SpG and hyABE8e‐SpRY) constructs. The sequences encoding the evolved adenosine deaminase TadA8e and DBD were fused to three PAM‐less/free SpCas9 nickase variants, Cas9n‐NG, SpGn and SpRYn. bpNLS, bipartite nuclear localization signal; 39 aa and 32 aa, 39‐aa and 32‐aa linker peptides; D10A, D10A substitution in Cas9 nickases. (b) Base‐editing efficiencies of PhieABEs compared to basic ABE8es and ABE7.10‐NG at 17 NGN‐PAM targets in T0 rice plants. A bases edited to G are highlighted in red. The numbers of edited and total T0 plants are given in parentheses, and data with grey background indicate the highest efficiency for each target among editors. ‘a’ and ‘b’ indicate data from previous studies included for comparative purposes (Hua et al., 2019b; Zeng et al., 2020b).
