Figure 2.
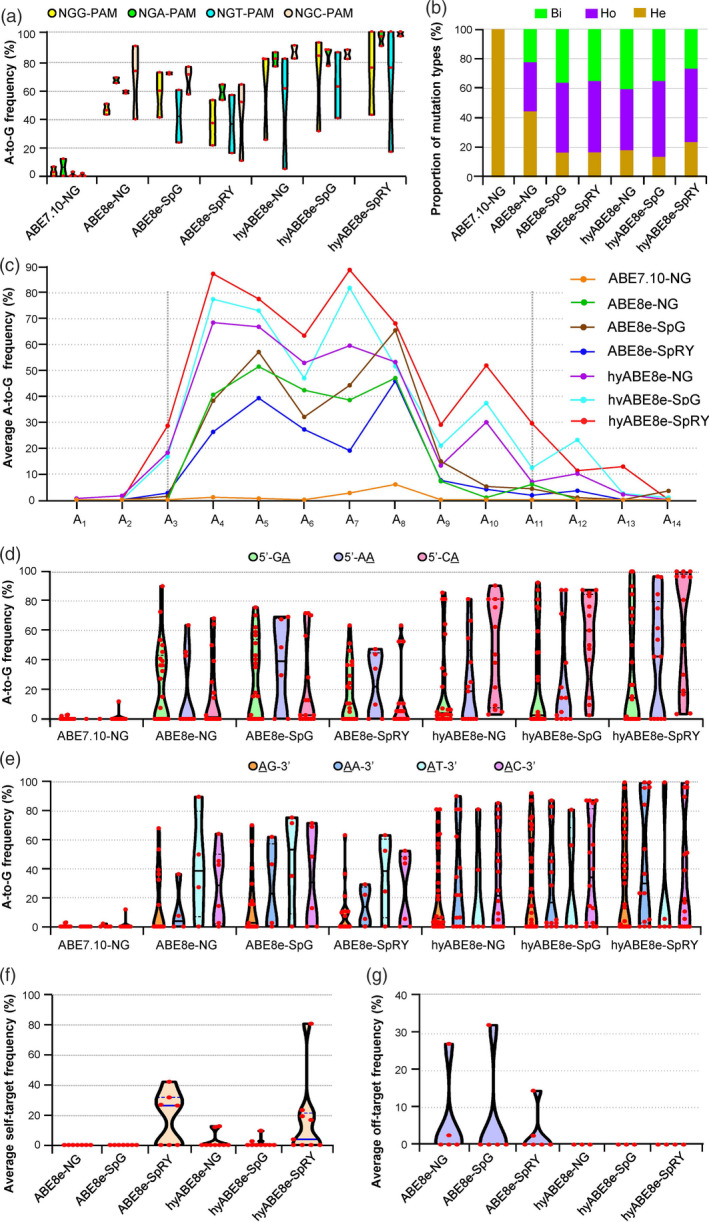
PhieABEs possess wider editing activity windows and better target sequence compatibility at NGN‐PAM sites. (a) Editing efficiencies at NGG‐, NGA‐, NGT‐ and NGC‐PAM targets in T0 plants with basic ABE8es and PhieABEs. ABE7.10‐NG (Hua et al., 2019b; Zeng et al., 2020b) was also used for comparison. (b) Proportion of mutation types induced by basic ABE8es and PhieABEs at all edited NGN‐PAM target sites (TS1–TS17). Bi, bi‐allelic mutations; Ho, homozygous; He, heterozygous. (c) Editing activity windows and efficiencies of PhieABEs, basic ABE8es and ABE7.10‐NG at TS1–TS17 sites. (d) and (e) Site preference analysis of 5’‐GA, 5’‐AA and 5’‐CA (d), and AG‐3’, AA‐3’, AT‐3’ and AC‐3’ (e) contexts for TS1–TS17 targets within the editing window A1–A14. (f) Self‐targeted editing efficiencies in the sgRNA expression cassettes at the TS1–TS17 sites. The hyABE8e‐SpRY shows weaker self‐editing activity than ABE8e‐SpRY in the T0 plants. (g) Off‐target editing frequencies of basic ABE8es and PhieABEs at sites homologous to TS1–TS4 and TS10. We observed no off‐target effects in T0 plants edited by PhieABEs.
