Figure 3.
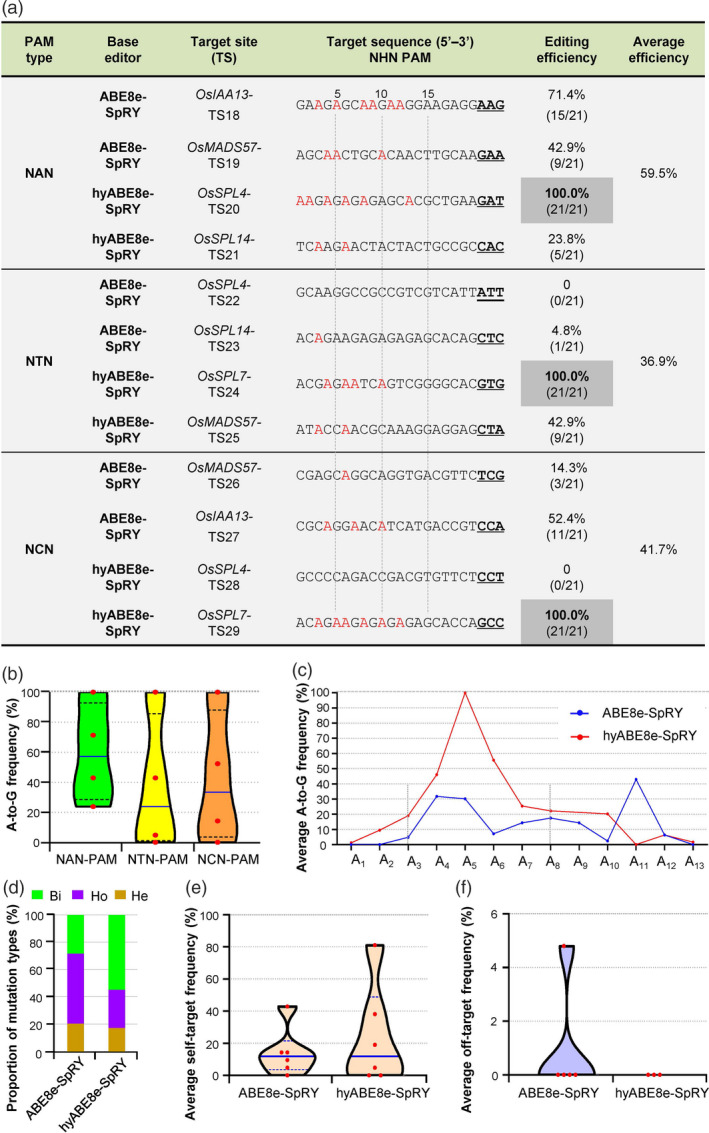
SpRY‐guided TadA8e allows efficient A‐to‐G editing at NHN‐PAM sites in rice. (a) Base‐editing efficiencies of ABE8e‐SpRY and hyABE8e‐SpRY at 12 NHN‐PAM (where H is A, T or C) sites (TS18–TS29) in T0 rice plants. A bases edited to G are highlighted in red. The number of edited and total T0 plants is given in parentheses. (b) Median editing efficiencies at NAN‐, NTN‐ and NCN‐PAM targets in the T0 plants edited by ABE8e‐SpRY or hyABE8e‐SpRY. (c) Editing activity windows and efficiencies of ABE8e‐SpRY and hyABE8e‐SpRY at the TS18–TS29 sites. (d) Proportion of mutation types induced by ABE8e‐SpRY and hyABE8e‐SpRY at the TS18–TS29 sites. (e) Self‐targeted editing efficiencies of the sgRNA expression cassettes at the TS18–TS29 sites. Both ABE8e‐SpRY and hyABE8e‐SpRY showed obvious self‐editing activities in the T0 plants. (f) Frequency of off‐target mutations at sites homologous to TS19, TS20, TS23, TS26 and TS29 induced by ABE8e‐SpRY or hyABE8e‐SpRY. No off‐target effect was detected in T0 plants edited by hyABE8e‐SpRY.
